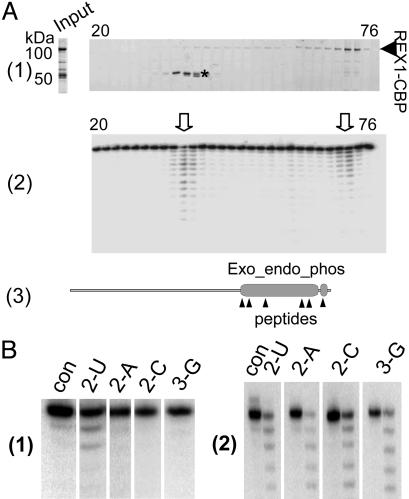
Fig. 2.
The 3′–5′ exonuclease activity of recombinant L. major LC-2. (A) The L. major LC-2–TAP fusion protein was expressed in a baculovirus system and affinity purified. The rLC-2–CBP was further purified over a HiTrapSP-HP column developed with a 0–2 M NaCl gradient over 60 ml. (A1) The fractions were analyzed on 8–16% SDS polyacrylamide gels that were stained with SYPRO Ruby. Input, the TAP-isolated material loaded on the column. The bands migrating at ≈50 kDa (*) were identified by mass spectrometry as L. major LC-2 degradation products. The two bands gave almost identical mass spectra, and all identified peptides from these bands corresponded to the C-terminal region of the LC-2–CBP fusion protein. (A2) The fractions were incubated with 5′ end-labeled 6-U RNA substrate, and the products were analyzed on a 7M urea/15% polyacrylamide gel. Two major peaks of exonuclease activity were observed (arrowheads). (A3) Diagram of the LC-2 protein showing the location of the Exo_endo_phos and CBP motifs and the sequenced peptides from the 50-kDa bands. (B1) U specificity of 100-kDa fraction (con, no enzyme control). The single-stranded RNAs used ending in 2 Us, 2 As, 2 Cs, or 2 Gs are indicated above each lane. (B2) Lack of U specificity of the 50-kDa REX1 fraction (con, no enzyme control for each digestion). The single-stranded RNAs used are indicated above each lane.