Fig. 12.
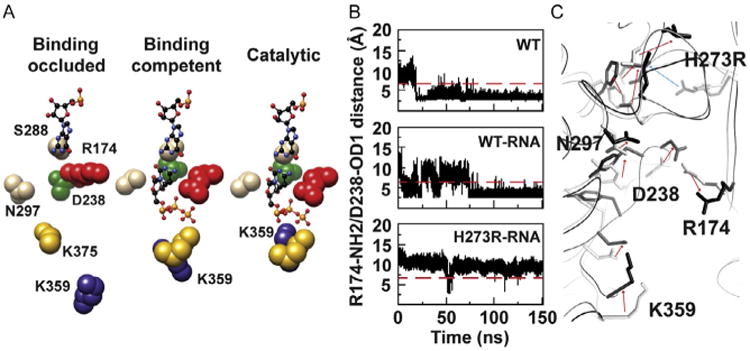
Physical mechanism of PV RdRp fidelity: an hypothesis. (A) Active-site conformational changes revealed by MD simulations of PV RdRp–RNA binary complex. (B) Time evolution of the distance between NH2 of Arg-174 and OD1 of Asp-238 atoms during 150 ns MD simulations of the free WT and the binary complexes of WT and H273R RdRps. (C) The average structures of WT-RNA (gray) and H273R-RNA (black) observed during the MD simulations are superimposed. Direction of arrows indicates the change from WT-RNA to H273R-RNA. Adapted from I.M. Moustafa, V.K. Korboukh, J.J. Arnold, E.D. Smidansky, L.L. Marcotte, D.W. Gohara, X. Yang, M.A. Sanchez-Farran, D. Filman, J.K. Maranas, et al., Structural dynamics as a contributor to error-prone replication by an RNA-dependent RNA polymerase, J. Biol. Chem. 289 (52) (2014) 36229–36248.
