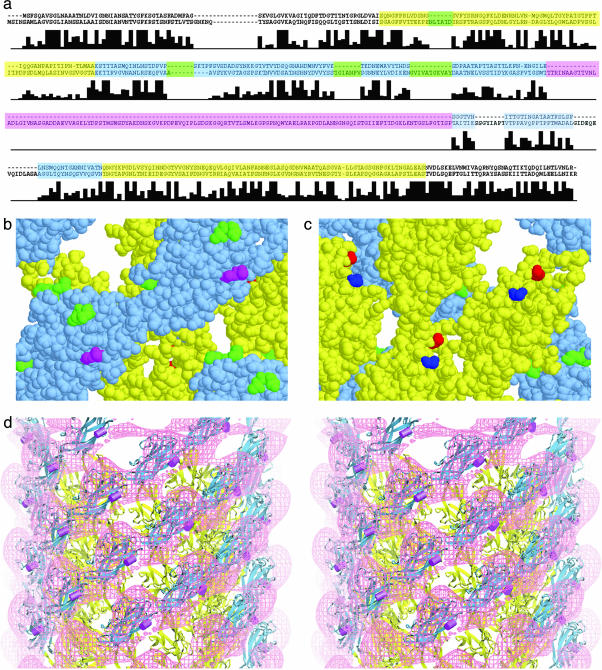
Fig. 4.
A comparison of the sequences of hook proteins. (a) Shown are the sequences corresponding to the proteins FlgE of S. typhimurium (upper sequence) and C. crescentus (lower sequence). The residues corresponding to domain D1 (or A in Fig. 3a) are shown in yellow, and those corresponding to domain D2 (or B in Fig. 3a) are shown in blue. The large insert found in C. crescentus but not S. typhimurium is shown in magenta. The other four inserts mapped in b and c are shown in green. Below the sequences is a bar graph showing sequence identity in FlgE for 13 bacterial species. (The height of the tallest bar corresponds to 13.) Note that the sequence assigned to domain D2 has more inserts than that of D1. (b) Space-filling model of hook subunits as seen from the outside. The sites of the small inserts are shown in green, and the site of the large insert in the hook of C. crescentus is shown in magenta. The yellow and blue portions correspond to domains D1 and D2, respectively, and the yellow and blue sequences in a.(c) Space-filling model of the hook subunits as seen from the inside the hook. The N and C termini, shown in blue and red, respectively, lie close to one another. The axis of the hook is vertical with the cell-proximal side at the bottom. (d) A stereo pair showing a surface representation of the difference map in which the S. typhimurium map is subtracted from the C. crescentus map. The atomic model derived by docking is shown with the site of the insert in C. crescentus marked by a magenta rod.