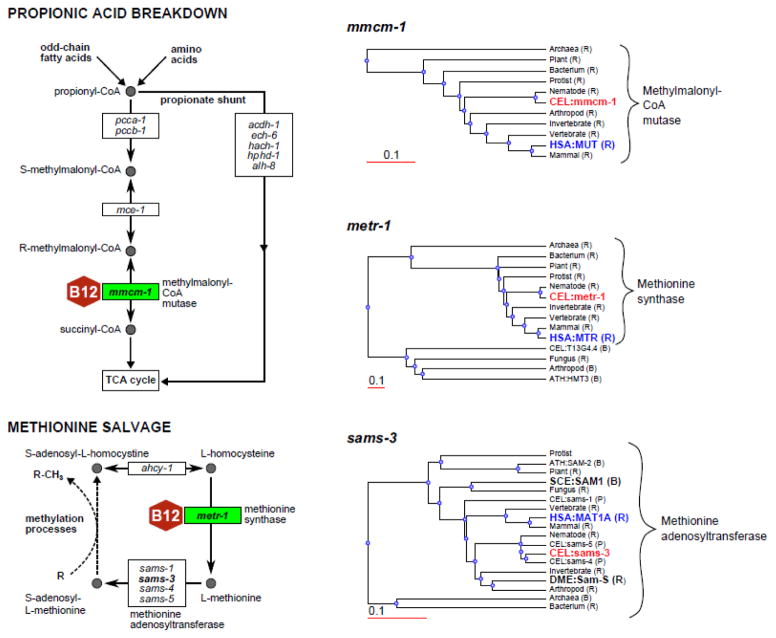
Figure 3.
C. elegans is an adequate model to study vitamin B12 metabolism but S. cerevisiae and D. melanogaster are not. Left panel shows two vitamin B12-dependent pathways. C. elegans genes encoding the enzymes are indicated. Recently discovered propionate shunt is also drawn without the details. For the genes in bold font, phylogenetic protein sequence trees are provided on the right panel. Trees were obtained from WormFlux (http://wormflux.umassmed.edu/) and edited for clarity. All reasonable best hits (B) and reciprocal best hits (R) (if available) from human (HSA), S. cerevisiae (SCE), and D. melanogaster (DME) were included in these trees [58]. Thus, S. cerevisiae (SCE) and D. melanogaster (DME) do not have the vitamin B12 enzyme homologs but they have an ortholog for methionine adenosyltransferase. Hits from A. thaliana (ATH) and C. elegans paralogs (P) are also shown. Identities of other organism genes are hidden except for the taxonomy.