Figure 1. Proteomic workflow for discovery and validation of novel T-bet phosphorylation sites.
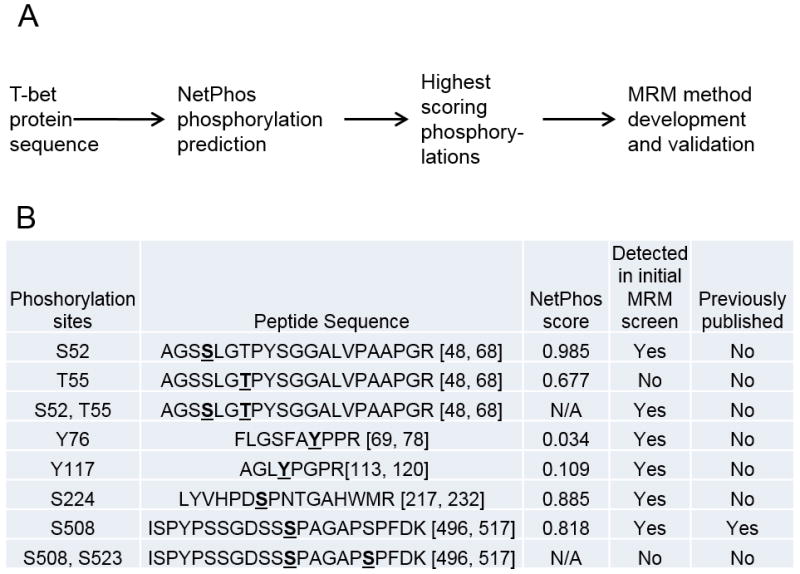
A, Mouse T-bet protein sequence was used as an input for phosphorylation site prediction analysis using NetPhos program. T-bet phosphorylations with the highest predicted scores were chosen for LC-MRM-MS method development. B, Phosphorylation sites and their NetPhos scores that were chosen for preliminary MRM method development.
