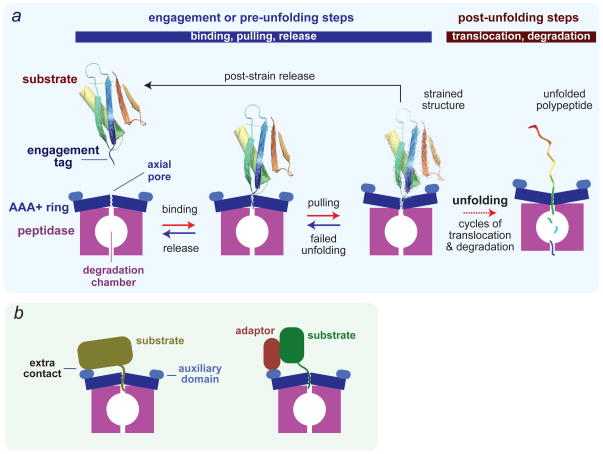
Figure 2. Substrate recognition and degradation.
(a) Minimal model for recognition, unfolding, translocation and degradation of a single-domain protein by a AAA+ protease. Reaction steps in the forward direction are ATP dependent. In the initial recognition step, a disordered engagement tag in the native protein substrate is bound in the axial pore of the AAA+ ring hexamer. ATP-fuelled conformational changes in the ring then pull on the substrate, which can result either in failed unfolding, substrate release or substrate denaturation. The probability of each of these outcomes depends on substrate stability, as a very stable protein might be bound and released many times resulting in unproductive hydrolysis of a substantial amount of ATP. Following forced unfolding, the denatured polypeptide is processively translocated through the pore and into the peptidase chamber for degradation. (b) Efficient recognition of some protein substrates requires secondary recognition signals, which are substrate sequences that bind to the AAA+ enzyme either directly or via adaptor proteins. In principle, these secondary signals might affect any of the pre-unfolding steps shown in panel a.