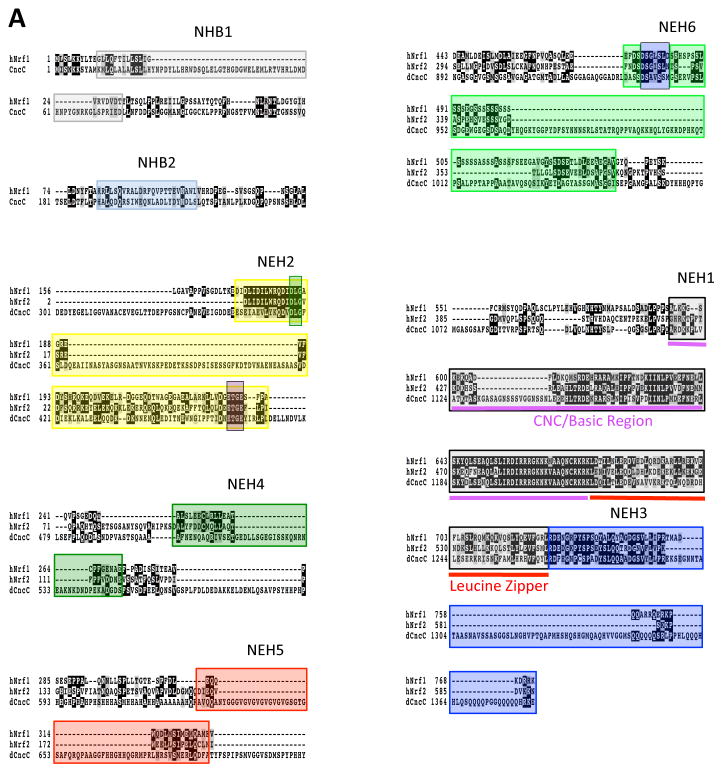
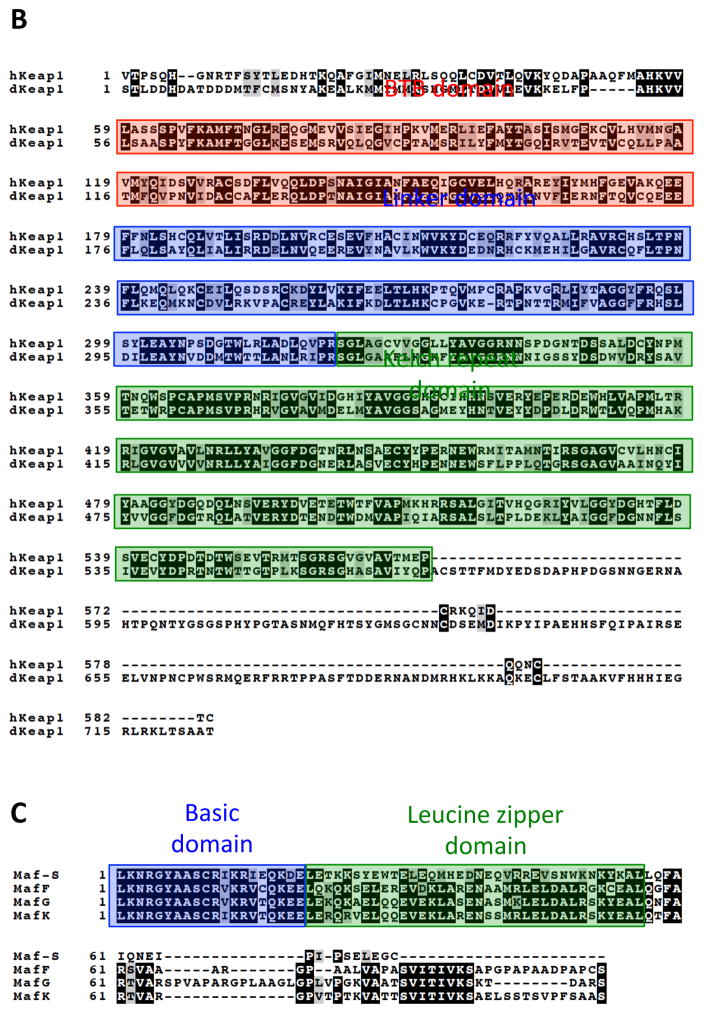
Figure 3. Amino acid sequence homology between Cnc, Keap1 and small Maf proteins of human and Drosophila.
A) Alignment of key functional elements in Nrf1, Nrf2, and CncC. Multisequence alignment using MAFFT, T-COFFEE, and BOXSHADE were used to compare publicly available amino acid sequences of human Nrf1, Nrf2, and CncC (McWilliam et al., 2013). Residues that are conserved between at least two of the sequences are shaded in black and residues with similar characteristics are shaded in gray. Domains and features described in Figure 1 are highlighted.
B) Alignment of Human and Drosophila Keap1. Multisequence alignment using MAFFT, T-COFFEE, and BOXSHADE were used to compare publicly available amino acid sequences of human Keap1 (hKeap1) and Drosophila Keap1 (dKeap1) (McWilliam et al., 2013). Residues that are conserved are shaded in black and residues with similar characteristics are shaded in gray. The BTB (Broad complex, Tramtrack, and Bric-a-Brac) domain contains key cysteine residues important for sensing oxidative stress (Zhang and Hannink, 2003). The linker domain plays a role in targeting Nrf2 for ubiquitination leading to its degradation as well as maintaining localization of the Nrf2-Keap1 complex in the cytoplasm (Zhang and Hannink, 2003). The Kelch repeat domain is responsible for Nrf2 interaction by interacting with the NEH2 regulatory domain of Nrf2 (Itoh et al., 1999).
C) Alignment of Mouse and Drosophila small Maf proteins. Multisequence alignment using MAFFT, T-COFFEE, and BOXSHADE were used to compare publicly available amino acid sequences of human Keap1 and Drosophila Keap1 (McWilliam et al., 2013). Residues that are conserved are shaded in black and residues with similar characteristics are shaded in gray. The basic-leucine zipper region is highlighted.