Figure 1.
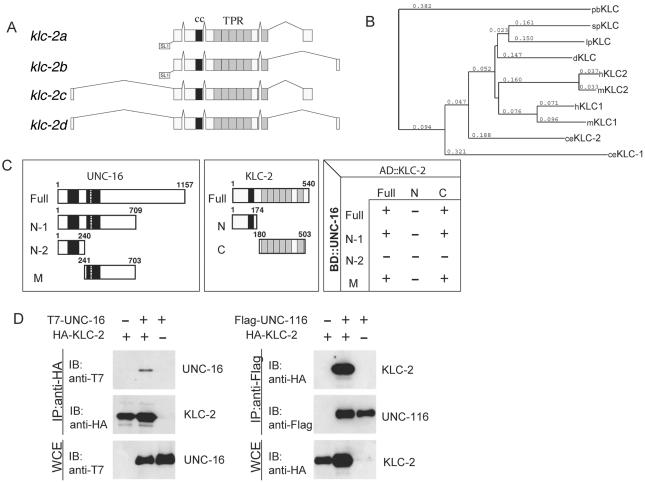
KLC-2 binds UNC-16 and the UNC-116 KHC. (A) Structures of klc-2 isoforms. The deduced four alternative splicing forms of klc-2 are shown. The black and gray boxes indicate the regions encoding the coiled-coil domain and TPR motifs, respectively. The trans-splicing sites are also indicated. (B) Alignment dendrogram generated using the neighbor-joining method within CLUSTALX. Numeric values indicate branch lengths in terms of percent divergence. pb, Plectonema boryanum (cyanobacterium); sp, Strongylocentrotus purpuratus (sea urchin); lp, Loligo pealii (squid); d, Drosophila; h, human; m, mouse; and ce, C. elegans. (C) Interaction between KLC-2 and UNC-16 in the yeast two-hybrid system. Various regions of UNC-16 fused with the LexA DNA-binding domain (BD) are shown on the left. The regions of KLC-2 fused with the GAL4 activation domain (AD) are shown in the middle. The plus (+) and minus (-) indicate positive and negative interactions between the various regions of UNC-16 and KLC-2 in the chart on the right. (D) Association of KLC-2 with UNC-16 and UNC-116. HEK 293 cells were transfected with control vector (-), HA-KLC-2, T7-UNC-16, and Flag-UNC-116 as indicated. Cell lysates were immunoprecipitated (IP) with anti-HA (left) and anti-Flag (right) antibodies. Immunoprecipitates were immunoblotted (IB) with anti-T7 (left panel) and anti-HA (right) antibodies. The amounts of immunoprecipitated HA-KLC-2 and Flag-UNC-116 were determined with anti-HA (left) and anti-Flag (right) antibodies, respectively. Whole-cell extracts (WCE) were immunoblotted with anti-T7 (left) and anti-HA (right) antibodies to determine total amounts of T7-UNC-16 and HA-KLC-2, respectively.
