Figure 6.
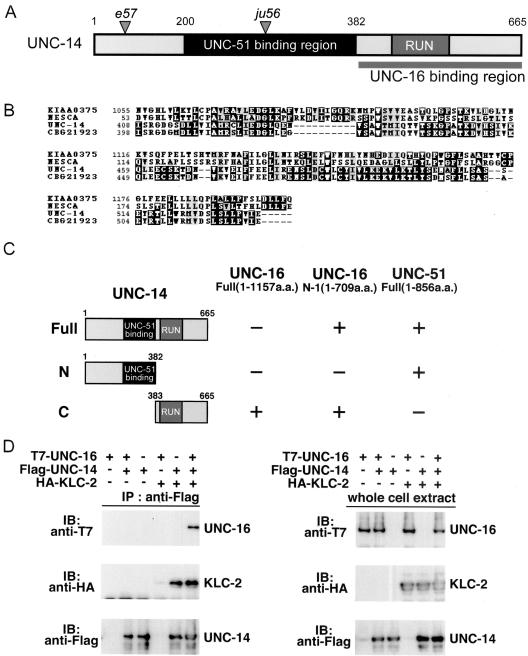
UNC-16 binds to the RUN domain region of UNC-14. (A) UNC-14 protein domain structure. The RUN domain and the UNC-16 binding region are shown; the shaded region shows the region that was previously reported to bind UNC-51 (Ogura et al., 1997). Positions of the e57 and ju56 early stop mutations are also indicated. (B) RUN domain sequence alignment. Multiple sequence alignment with human NESCA and conceptual translation of KIAA0375, C. elegans UNC-14, and C. briggsae CBG21923 RUN domains constructed with CLUSTALW and displayed by BOXSHADE. (C) Interaction of UNC-14 with UNC-16 and UNC-51 in the yeast two-hybrid system. UNC-16 N-1 and UNC-51 were constructed with the LexA DNA-binding domain (BD). Various regions of UNC-14 constructed with the GAL4 activation domain (AD) are shown on the left. Plus (+) and minus (-) indicate positive and negative interactions, respectively. (D) Coimmunopreciptation of UNC-16 and KLC-2 with UNC-14. HEK 293 cells were transfected with control vector (-), T7-UNC-16, Flag-UNC-14, and HA-KLC-2, as indicated. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody (left panels). Immunoprecipitates were immunoblotted (IB) with anti-T7 (top panel) and anti-HA (middle panel) antibodies. The amounts of immunoprecipitated Flag-UNC-14 were determined with anti-Flag antibody (bottom panel). Whole-cell extracts were immunoblotted with anti-T7, anti-HA, and anti-Flag antibodies to determine total amounts of T7-UNC-16, HA-KLC-2, and Flag-UNC-14, respectively (right panels).
