Figure 8.
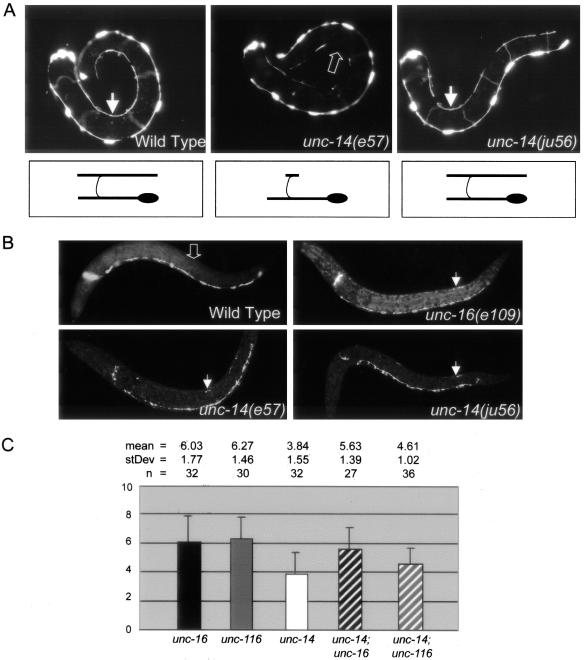
unc-14 regulates synaptic vesicle localization and functions in the same pathway as unc-16 and unc-116. (A) DD cell morphology. Panels show L1 stage animals expressing Punc-25-GFP (juIs76) to visualize the morphology of the DD motor neurons. In wild-type animals, six DD cell bodies reside along the ventral nerve cord. Each extends a process along the ventral nerve cord, around the side of the body (commissures out of the focal plane), and along the dorsal nerve cord. The arrows in the left and right panels indicate fully extended dorsal DD processes in wild-type and unc-14(ju56) L1 animals. The open arrow in the middle panel indicates missing dorsal DD processes in an unc-14(e57) L1 animal. Drawings below the animals demonstrate the morphology of an individual DD motor neuron representative of each genotype. (B) SNB-1::GFP (juIs1) localization in L1 animals. Although wild-type L1s have no SNB-1::GFP along their dorsal DD processes (open arrow in top left panel), SNB-1::GFP is misaccumulated along the dorsal DD processes of unc-16 and unc-14 mutant L1 animals (closed arrows in top right and bottom panels). (C) Quantification of L1 dorsal SNB-1::GFP phenotype. The extent of dorsal SNB-1::GFP was scored in L1 animals on a scale from 0 to 25 (see Materials and Methods) in unc-16(e109), unc-116(e2281), and unc-14(ju56) single mutants, and in unc-14(ju56); unc-16(e109) and unc-14(ju56); unc-116(e2281) double-mutant animals. The mean, SD, and n are indicated above the graph. By Student's t-tests (2 tail/unpaired/unequal variance), the L1 SNB-1::GFP phenotypes of unc-16(e109) single mutants and unc-16(e109); unc-14(ju56) double mutants are not significantly different (p = 0.333), and the L1 SNB-1::GFP phenotypes of unc-14(ju56) single mutants and unc-14(ju56); unc-116(e2281) double mutants are not significantly different (p = 0.21). Although other pair combinations have significantly different mean scores, all differences are less than additive. p-values for other pair-wise comparisons are as follows: unc-16(e109) and unc-14(ju56) (p = 1.9e-6), unc-14(ju56) and unc-14(ju56); unc-16(e109) (p = 1.9e-5), unc-116(e2281) and unc-14(ju56) (p = 3.3e-8), and unc-116(e2281) and unc-14(ju56); unc-116(e2281) (p = 3.2e-6).
