Figure 8.
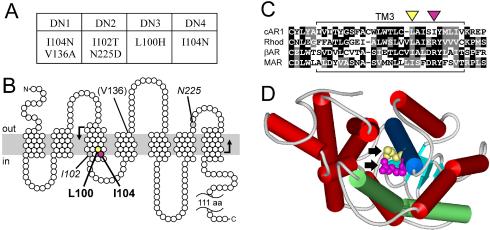
Activating mutations are clustered in a conserved region of the third transmembrane helix. (A) Summary of mutations present in cAR1 mutants DN1-4. (B) Topological model of cAR1 (based on Klein et al., 1988) indicating the amino acids mutated in DN1-4. Those causing dominant receptor activation are shown in bold, those whose role is ambiguous are italicized, and those unlikely to have a role are in parentheses. The plasma membrane is depicted as a gray bar. The region mutagenized is bounded by bent arrows. (C) Amino acid sequence alignment of the third transmembrane domains of cAR1 (residues 79-113), bovine rhodopsin (residues 110-144), bovine β-adrenergic receptor (residues 131-165), and human M1 muscarinic ACh receptor (residues 98-132). Black and gray boxes emphasize identical and similar residues, respectively. Hyphens indicate gaps introduced for optimal alignment. Yellow and magenta triangles identify L100 and I104 in cAR1, respectively. (D) Rhodopsin's structure (Palczewski et al., 2000) viewed from cytoplasm, highlighting TM3 (blue) and TM6 (green). Arrows indicate residues corresponding to L100 (yellow) and I104 (magenta) of cAR1. WebLab ViewerLite software (Accelrys, San Diego, CA) was used for structure modeling.
