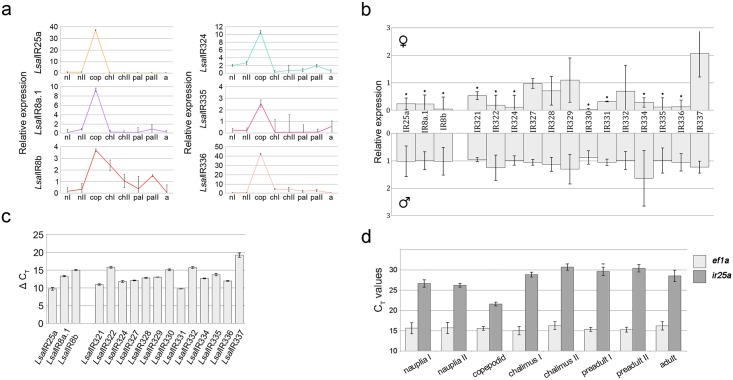
Fig 2. IRs expression levels through L. salmonis life cycle and in selected tissues in adult lice.
(a). Relative expression level of three co-receptors and selected antennal IR genes in all developmental stages. The highest expression is in planktonic copepodids for all tested genes. Quantification was performed on batches of 100 nauplia I, nauplia II, copepodids, 20 chalimus I, 10 chalimus II, 4 preadult I females, 2 preadult II females, 1 adult female, 10 preadult I males, 4 preadult II males and 1 adult male (n = 5). Error bars show standard deviation. Abbreviations: nI—nauplia I, nII—nauplia II, cop—copepodid, chI—chalimus I, chII—chalimus II, paI—preadult I (both sexes), paII—preadult II (both sexes), a–adult (both sexes). (b). Comparison of transcripts level of antennal IRs in adults of both sexes tested by qRT-PCR. All co-receptors and 8 out of 13 antennal genes reveal higher expression in males than in females. Expression-PCR was performed on 1 adult female or male (n = 5). Error bars show standard deviation. Each louse was analysed separately and standard deviations represent individual differences. Asterisks indicate statistically significant differences in expression level between male and female (p < 0.05). Statistical analysis was performed using Independent-Samples T-Test. Comparison was performed for each gene independently. (c). Transcript level of IR genes in the copepodid stage. mRNA level for each tested gene is presented as a ΔCT value. Expression-PCR was performed on batches of 100 copepodids (n = 5). Error bars show standard deviation. (d). Comparison of CT values of LsIR25a and reference gene LsEF1A in all developmental stages. LsEF1A is expressed at the same level in all developmental stages whereas the CT values for LsIR25a vary considerably and are the lowest in the copepodid stage. Q-PCR were done on same samples as in Fig 2a (n = 6).