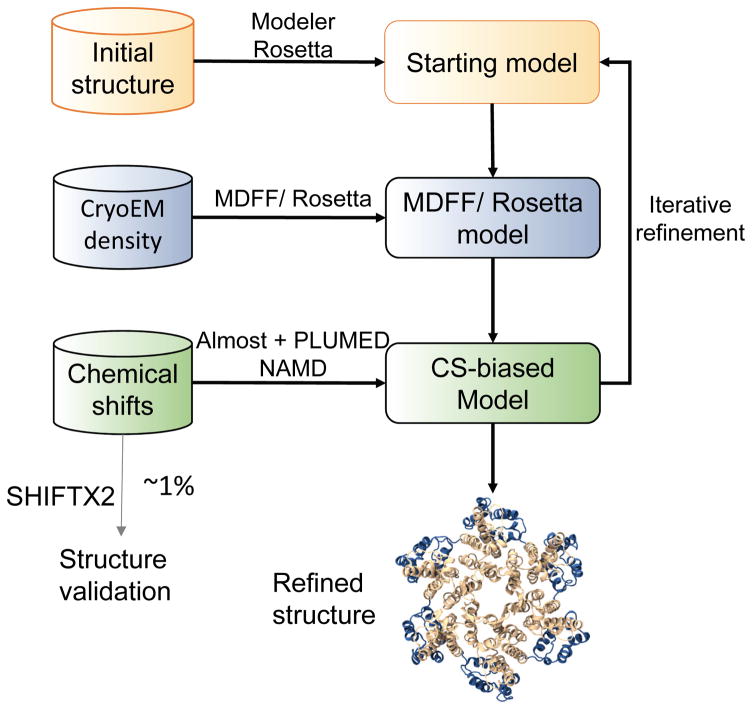
Figure 1.
Schematic diagram illustrating NMR CS-biased cryoEM structure refinement. A starting model is fit into the experimental cryoEM density map (4–6 Å resolution) using molecular dynamics flexible fitting (MDFF). The predicted chemical shifts (CSs) for the resulting model are compared with experimental NMR CSs, and the differences between these are incorporated as a biasing potential in MD, such as NAMD. The biasing forces are calculated using Almost and send to NAMD using PLUMED. The resulting CS-biased model is then iteratively refined. In parallel, CSs calculated from the resulting model using SHIFTX2 are compared to a small subset of experimental NMR CSs that have not been used during the CS-biased model refinement procedure for model validation.