Figure 6.
Prospective Isolation of Early-Stage Naive Cells
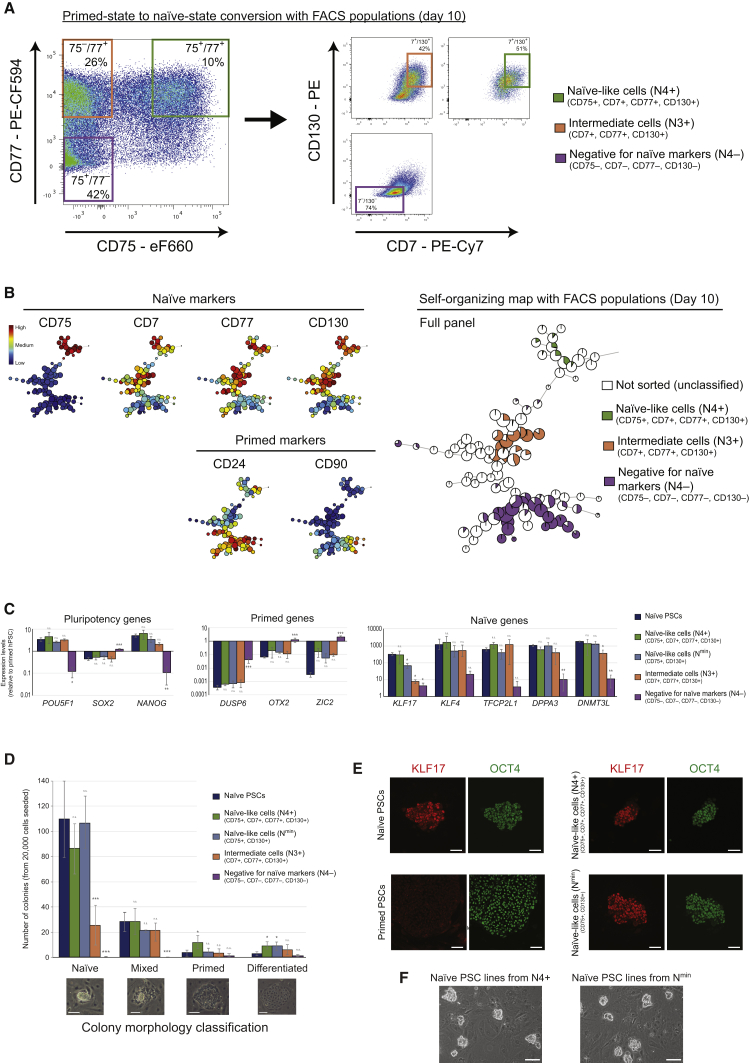
(A) Flow cytometry dotplots of day 10 cells during primed-state to naive-state conversion of H9 PSCs under t2i/L+PKCi conditions. Left: the levels of two naive-specific markers, CD75 and CD77. Based on unstained, live, human day 10 samples, three cell sorting gates have been drawn that correspond to CD75+/CD77+ (green box), CD75–/CD77+ (orange box), and CD75–/CD77– (purple box) cell populations. Right: the levels of CD7 and CD130 proteins for the same three gated cell populations. Boxed areas indicate the N4+ (green), N3+ (orange), and N4– (purple) cell populations that were used for subsequent experiments. The percentage of cells within each cell sorting gate relative to all live, human cells is shown. Note that the values do not take into account additional gates; for example, to exclude primed-state markers. See Figure S5C for the Nmin gating strategy.
(B) FlowSOM visualization of the flow cytometry data for day 10 cells during primed to naive conversion. The minimal spanning tree of the self-organizing map displays an unsupervised clustering of the sample based on the cell surface protein expression levels (right). The cells corresponding to each cell sorting population, N4+, N3+, and N4–, are indicated. The heatmap shows the expression level of each cell surface protein marker in the cell clusters (left). See Figure S5A for FlowSOM visualization of WIBR3 PSCs on day 10 of primed state-to-naive state conversion and Figure S5D for FlowSOM visualization of Nmin cells.
(C) qRT-PCR analysis of gene expression levels in the different cell-sorted populations and established naive PSCs. Expression levels are shown on a log scale relative to primed PSCs. Data show the mean ± SD of three or four biological replicates and were compared to established naive PSCs using an ANOVA with Dunnett’s multiple comparisons test (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005).
(D) Scoring of colony morphology after transferring the different cell-sorted populations into naive PSC conditions. Colonies were categorized as naive, mixed, primed, and differentiated; examples are shown below. Data show the mean ± SD of three or four biological replicates and were compared to established naive PSCs using an ANOVA with Dunnett’s multiple comparisons test (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005). Scale bars, 100 μm.
(E) Immunofluorescence microscopy for KLF17 (a naive-specific protein) and OCT4 (a protein expressed by naive and primed PSCs) reveals that N4+ and Nmin cell-sorted populations can generate KLF17+/OCT4+ colonies that are similar to established naive PSCs. Scale bars, 100 μm.
(F) Phase contrast images showing representative fields of view of N4+ and Nmin cell-sorted populations that have been propagated under t2i/L+PKCi naive PSC conditions for three passages. Scale bars, 100 μm. See Figure S5B for similar results using WIBR3 PSCs under 5i/L/FA conditions.