Figure 1.
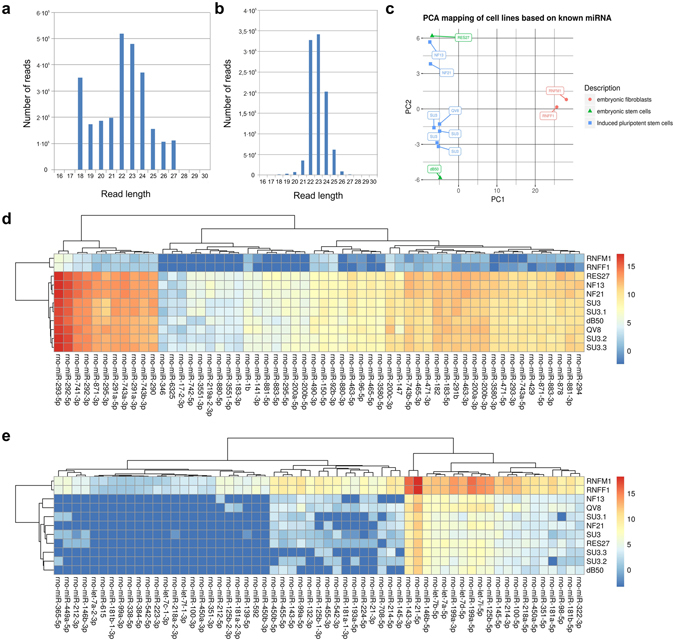
(a) The number of informative reads at different lengths for sample SU3. (b) The number of informative reads at different lengths for sample SU3 that were mapped to known rat miRNAs. (c) Principal component analysis of 11 samples based on RPM expression of 219 differentially expressed miRbase miRNAs. (d and e) Heatmaps of 56 upregulated miRNAs in PSCs (d) and 59 upregulated miRNAs in EFs (e). Known miRNAs with an extremum fold change more than 10, and the log2 transformation.
