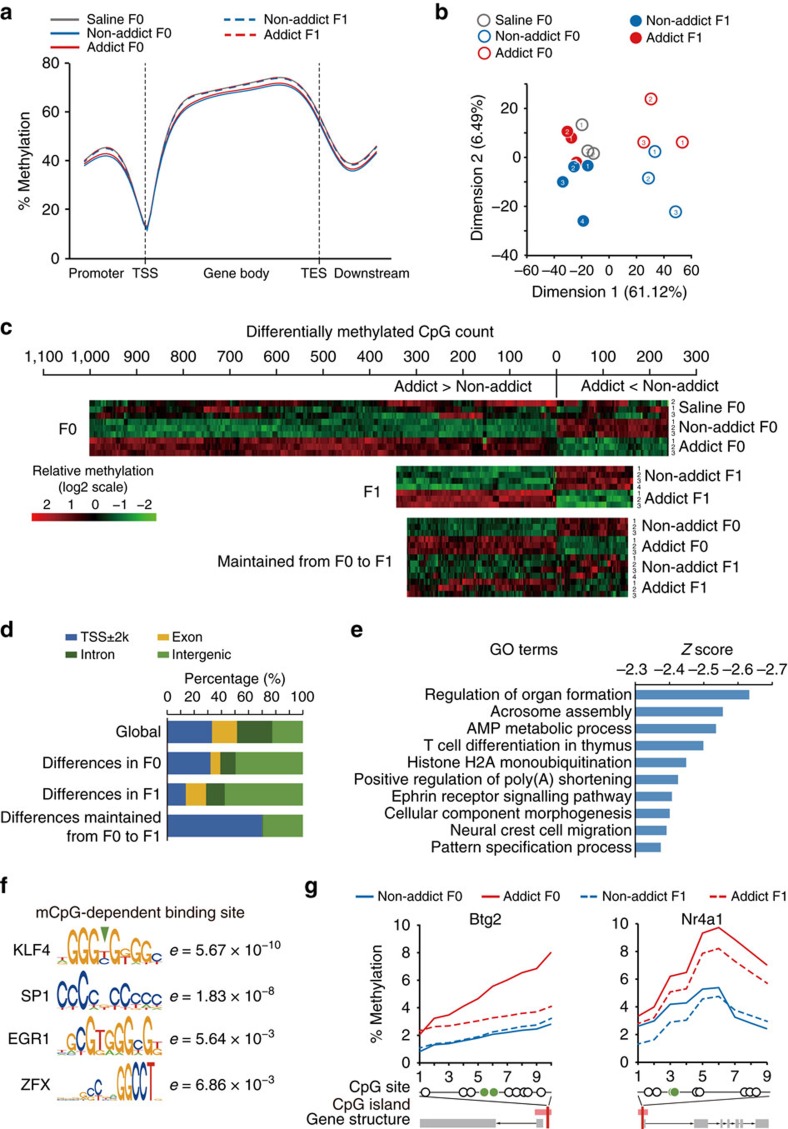
Figure 8. Identification of maintained DNA methylation alterations in the sperm of Addict F0 and F1 rats.
(a) The average methylation levels of CpG sites upstream of the transcription start site (TSS), within the gene body, and the downstream of transcription termination sites (TES) in all genes detected. Addict F0 and Non-addict F0 showed lower global methylation level than Saline F0. (b) Euclidean distance-based multidimensional scaling indicated segregation of generation and addiction state. Number in circle refers to subject number in each group, corresponding to the subject number in c. (c) Methylation in 1,244 CpGs was significantly different between Addict F0 and Non-addict F0, and 522 differentially methylated CpGs were detected in Addict F1 versus Non-addict F1. The differential methylation was maintained in 475 CpGs across F0 to F1 generation. FDR≤0.05 was considered significant. (d) CpG distributions in TSS±2,000 bp area, exons, introns, and other intergenic elements. CpG methylation differences maintained from F0 to F1 were enriched in TSS±2,000 bp area. Differences maintained from F0 to F1/Global, odds ratio (95% confidence intervals), 273.95 (55.82-1340.73). (e) Ontology analysis of CpGs (in TSS±2,000 bp region) with differential methylation maintained from F0 to F1. Z scores from Enrichr were acquired and ranked. (f) Motif enrichment analysis of TSS±2,000 bp region by rGADEM and MotIV. The results revealed specific enrichment of KLF4 binding sites (e=5.67 × 10−10). (g) Methylation profiles of representative CpGs within differential methylation region (DMR) of Klf4-regulated genes. DMRs were calculated by BiSeq. Group averages are plotted. Btg2: DMR, Chr13: 55970169–55970230, three-way RM ANOVA, Faddict × generation × site (9, 81)=7.863, P=3.03 × 10−8; Bonferroni post hoc, P=3.5 × 10−14, Non-addict versus Addict. Nr4a1: DMR, Chr7: 140712984–140713087; three-way RM ANOVA, Faddict × generation × site (8, 72)=0.832, P=0.58; Bonferroni post hoc, P=1.1 × 10−11, Non-addict versus Addict. Non-addict F0, n=3; Addict F0, n=3; Non-addict F1, n=4; Addict F1, n=3.