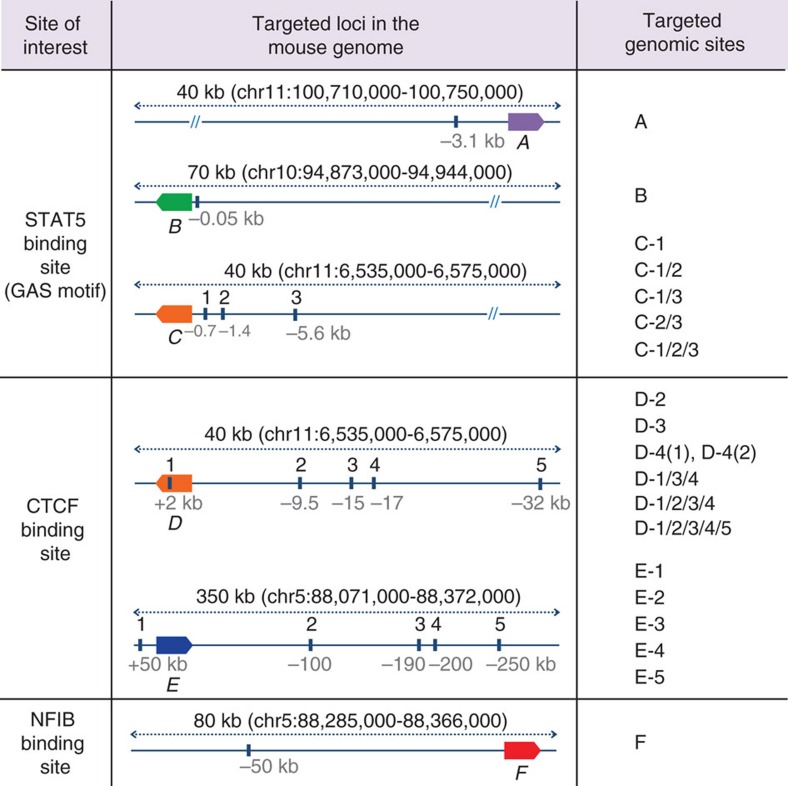
Figure 1. Targeting 17 sites in the mouse genome with CRISPR/Cas9.
STAT5 TF-binding sites (GAS motif), CTCF-binding regions and an NFIB-binding site were targeted for deletion. STAT5-binding sites in three gene loci (A, Stat5 (ref. 20); B, Socs2 (ref. 26); C, Wap19) were targeted individually (A, B and C-1). The three STAT5-binding sites in the Wap super-enhancer19 were deleted in four different combinations (C-1/2, C-1/3, C2/3 and C-1/2/3). Combined deletion of two or more STAT5-binding sites within the same gene locus was accomplished through a successive (two-steps) or simultaneous (one-step) targeting. A total of 11 CTCF-binding sites in two gene loci (D, Wap; E, Csn) were targeted individually and in different combinations. (F), an NFIB-binding site in the Csn locus was targeted (Supplementary Fig. 1). Target mutations were identified in 632 founder mice and 54 mutant lines were established. The positions of reference genes in the respective loci are indicated as coloured boxes. A, Stat5a; B, Socs2; C and D, Wap; E, Csn1s1; F, Csn3.