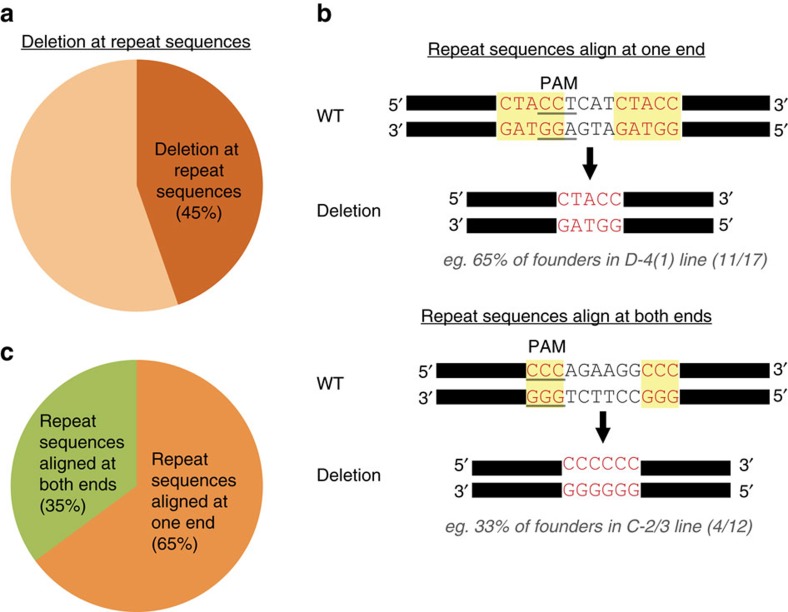
Figure 6. Preferential deletions at repeat sequences.
(a) Average frequency of deletions found at repeat sequences by single sgRNA injections. Repeat sequences were aligned in ∼45% at one end or both ends (total n=122; deletion at repeat sequences, n=56). Only deletions obtained from injections with single sgRNAs were analysed to avoid effects of multiple variables. Deletions obtained from single sgRNA injections: Deletions targeting TF-binding site B, C-1, C-2/3, D-2, D-3, D-4(1) and F. (b) Representative examples of repeat sequences aligned at one end (upper panel) and both ends (bottom panel). More than 60% of mutant founder mice targeting the genomic site D-4 exhibited the exact same deletion that retained only a single copy of repeat sequence. More than 30% of mutant founder mice targeting the genomic site C-2/3 showed the exact same deletion and repeat sequences remained at both ends. (c) Percentage of repeat sequences aligned at one end and both ends in founder mice carrying deletions at repeat sequences.