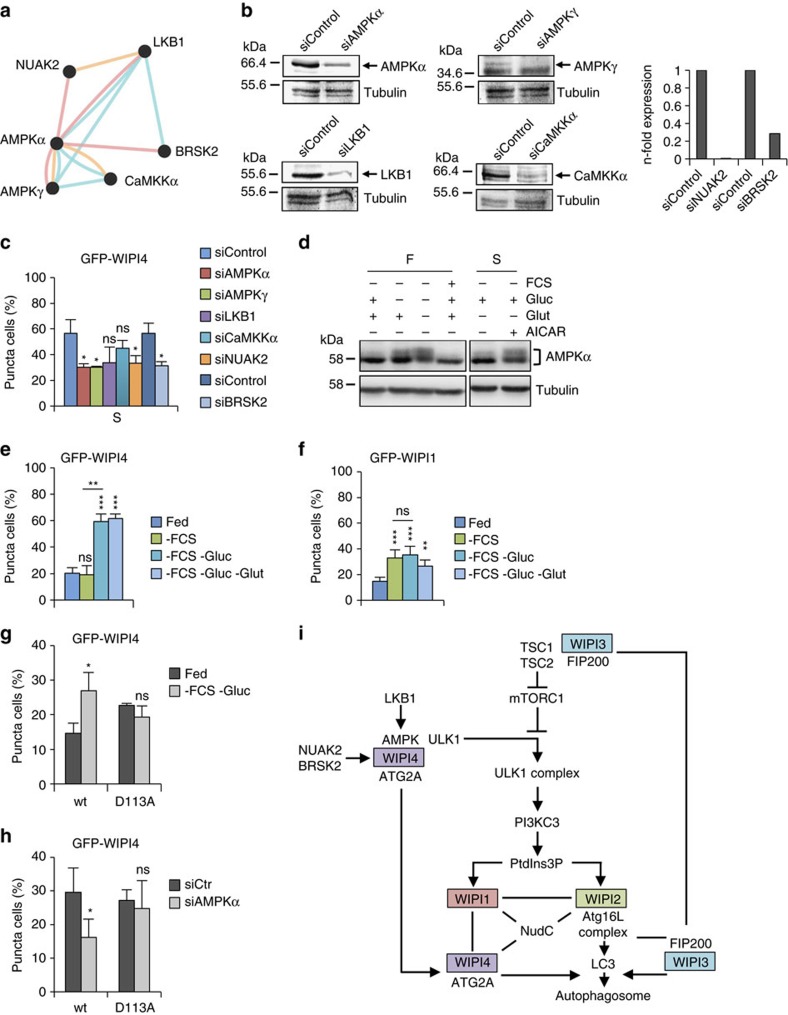
Figure 8. Glucose-starvation-mediated AMPK activation regulates WIPI4.
(a) A lentiviral-based shRNA screening approach targeting the human kinome used for the assessment of autophagy identified AMPK and the AMPK-related protein kinases NUAK2 and BRSK2, along with CaMKKα, as autophagy regulators (for details see Methods section and Supplementary Fig. 8). Reported (red) and predicted (yellow) proteins interactions, and pathway interactions (blue) are indicated (GeneMANIA). (b) Stable GFP-WIPI1 U2OS cells were transfected with siRNAs for AMPKα, AMPKγ, LKB1, CaMKKα, NUAK2 or BRSK2 and knock-down confirmed by immunoblotting (left panels) or quantitative RT–PCR (right panels). (c) Stable GFP-WIPI4 U2OS cells with siRNAs targeting AMPKα, AMPKγ, LKB1, CaMKKα, NUAK2 or BRSK2 were starved (S) for 3 h. Mean percentages of GFP-WIPI4-puncta-positive cells (300 cells per condition, n=3) are presented. (d) U2OS cells were fed (F) or starved (S) with or without glucose, glutamine or AICAR and immunoblotted using anti-AMPKα and anti-tubulin antibodies. Stable GFP-WIPI4 U2OS cells (e) or GFP-WIPI1 (f) were fed or treated with complete medium without FCS (−FCS), without glucose (−FCS −Gluc) or without glucose and glutamine (−FCS −Gluc −Glut). Mean percentages of GFP-WIPI4-puncta-positive cells (300 cells per condition, n=3) (e) and GFP-WIPI1-puncta-positive cells (up to 3,904 cells per condition, n=6) (f) are presented. (g) U2OS cells expressing GFP-WIPI4 WT or GFP-WIPI4 D113A mutant were fed or glucose-starved (−FCS −Gluc) for 3 h. Mean percentages of GFP-WIPI4-puncta-positive cells (300 cells per condition, n=3) are presented. (h) U2OS cells expressing GFP-WIPI4 WT or GFP-WIPI4 D113A mutant with siControl (siCtr) or siAMPKα were glucose-starved (−FCS −Gluc) for 3 h. Mean percentages of GFP-WIPI4-puncta-positive cells (500 cells per condition, n=5) are presented. (i) A predicted model for the differential contributions of human WIPI β-propeller proteins in autophagy. Statistics and source data can be found in Supplementary Data 1. Mean±s.d.; heteroscedastic t-testing; P values: *P<0.05, **P<0.01, ***P<0.001, ns: not significant.