Figure 2.
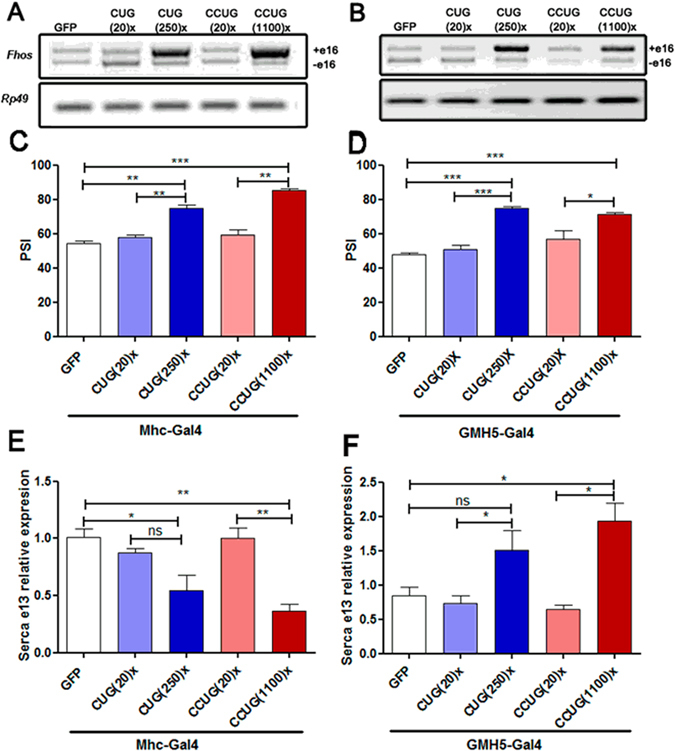
CUG and CCUG expansions cause Muscleblind-dependent missplicing. (A,B) Representative semi-quantitative RT-PCR showing inclusion of Fhos exon 16′ in flies expressing the indicated constructs in muscle (A) or heart (B) under the control of Mhc-Gal4 or GMH5-Gal4, respectively. Endogenous Rp49 was used for normalization. Percentage of exon 16′ inclusion, revealed that expression of long CUG or CCUG repeats in the fly muscle (C) or heart (D), favored increased use of this exon. qRT-PCR results of Serca exon 13 expression relative to Rp49 expression, confirmed that the use of this exon in the flies expressing the expanded repeats and the control flies is significantly different in muscle (E) and heart tissues (F). The histograms show the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
