Abstract
Background & objectives:
Slums are considered as hotspots of tuberculosis (TB). The study of genetic diversity and drug susceptibility profile of Mycobacterium tuberculosis (MTB) will help understand the transmission dynamics and can be used for better prevention and control of the disease. The aim of this study was to determine the drug susceptibility profiles and genetic diversity using the random amplified polymorphic DNA (RAPD) and mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU VNTR) of MTB isolates from sputum samples of pulmonary TB patients residing in the two slums of Jaipur city in Rajasthan, India.
Methods:
Sputum samples collected from pulmonary TB patients, their contacts and suspects during 2010-2012 were processed for microscopy and mycobacterial culture. Drug susceptibility testing was done by one per cent indirect proportion method on Lowenstein–Jensen medium for first-line anti-TB drugs rifampicin, isoniazid, ethambutol and streptomycin. MTB DNA was extracted by physicochemical method, and DNA fingerprinting was done by RAPD and MIRU VNTR analysis.
Results:
Among 175 sputum samples collected, 75 were positive (43.8%) for acid-fast bacilli, 83 for MTB culture and four were contaminated. Fifty two isolates (62.7%) were fully sensitive to four drugs, and five (6%) were multidrug resistant (MDR). RAPD analysis of 81 isolates revealed six clusters containing 23 (28.4%) isolates, and 58 (71.6%) were unique. MIRU VNTR analysis clustered 20 (24.7%) isolates, and 61 (75.3%) were unique.
Interpretation & conclusions:
About 62.7 per cent isolates from the sputum samples from slum areas were sensitive to four drugs; six per cent of isolates were MDR. Poly-resistance other than MDR was high (16%). About one-fourth isolates were clustered by either method. RAPD was rapid, less expensive but had low reproducibility. MIRU VNTR analysis could identify to greater extent the epidemiological link in the population studied.
Keywords: Drug susceptibility testing, mycobacterial interspersed repetitive units-variable number of tandem repeats, Mycobacterium tuberculosis, random amplified polymorphic DNA
India is one of the high tuberculosis (TB) prevalent nations in the world1. Several high TB incidence nations possess some hyperendemic ‘hotspots’ which contribute immensely to TB transmission2. Slums are one of these ‘hotspots’, characterized by poverty, overcrowding, concentration of lower socio-economic class and associated with multiple health problems, malnutrition, poorly ventilated houses and unhygienic environment3. High transmission of TB has been reported in communities characterized by dense population and poor hygienic conditions in resource-poor and high TB incidence countries like India4. Jaipur, the State capital of Rajasthan, has over 235 slum areas, in which around 4.87 lakh people live (22.4% of the total population)5. The major diseases reported in these areas are TB (3.0%), blindness (1.9%) and leprosy (0.8%)6. High prevalence of TB in slums has been reported from India7,8,9 and western countries2,3. High prevalence of multidrug-resistant (MDR) TB has been reported from slums in Dhaka10 and Mumbai11, but no such information is available from slums of Jaipur. The study of genetic diversity of Mycobacterium tuberculosis (MTB) strains by DNA fingerprinting in an area can provide insights on transmission dynamics11,12,13. IS6110-based restriction fragment length polymorphism (RFLP) DNA fingerprinting of MTB has been considered an established standard, but the major drawback is that it is lengthy, cumbersome and a significant proportion of Indian MTB isolates have low or zero copy numbers of these sequences which significantly reduces its utility in India14,15,16. This has necessitated the use of other rapid and reliable fingerprinting methods for the typing of MTB isolates such as random amplified polymorphic DNA (RAPD) fingerprinting13, mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU VNTR) analysis17 and spoligotyping18. Analysis of drug resistance patterns and genotypic diversity of the MTB isolates can provide useful information for understanding the transmision dynamics. The present study was thus planned to analyze the drug resistance profile and genotypic diversity of MTB isolates using two polymerase chain reaction (PCR)-based systems, i.e. IS 986F-based RAPD analysis and 12 loci-based MIRU VNTR analysis from sputum samples of pulmonary TB patients and their contacts residing in the two closely located slums (Eidgah & Nagtalai) of Jaipur city, India.
Material & Methods
The study was conducted during 2010-2012 at two urban adjacent slums of Jaipur city (Eidgah & Nagtalai) from the Revised National Tuberculosis Control Programme Directly Observed Treatment Short Course (RNTCP DOTS) centres of the area. The sample size was calculated using the formula n=t2 × p (1 − p)/m2, taking national prevalence of MDR in new smear positive (NSP) cases as 3 per cent (p), margin of error at 3 per cent (m), 95 per cent confidence interval (t standard value 1.96), resulting in the power of study being 0.95. The proposed sample size was calculated to be 124. In the present study, 171 TB cases were included, of whom 160 were NSP cases.
Inclusion & exclusion criteria: Consecutive, newly diagnosed pulmonary TB patients and pulmonary TB suspects (as per the RNTCP guidelines19) visiting the DOTS centre and residing in Eidgah and Nagtalai slums of Jaipur; known TB patients [less than two months of anti-tubercular therapy (ATT)]; MDR-TB suspects (treatment failures of Cat II regimen); symptomatic contacts suspected of TB of all the above; patients who consented to participate in the study and gave their written consent were included in the study. Known TB patients with greater than two months of ATT [whose samples would probably not yield positive growth on Lowenstein–Jensen (LJ) culture], as well as all patients/suspects/contacts not willing to participate in the study were excluded. Known HIV-positive patients were also excluded from the study.
Clinical details & sample collection: Detailed history of the patients which included age, sex, HIV status, and intake of ATT in the past was noted. Written informed consent was taken from all the patients/suspects enrolled in the study. The study protocol was approved by the Institutional Ethics Committee of the SMS Medical College, Jaipur. Early morning sputum samples (4-5 ml) were collected in sterile containers and transported to the Mycobacteriology Laboratory [RNTCP-approved culture & drug susceptibility testing (DST) Laboratory] of SMS Medical College, Jaipur, the same day.
Biosafety precautions: All procedures were performed in an RNTCP-accredited biosafety level II (BSL II) culture and DST Laboratory. All precautions were taken as per RNTCP guidelines19. This was also done for the sputum samples after testing as well as for culture isolates and media used for the study.
Smear microscopy & culture on Lowenstein–Jensen (LJ) medium: Sputum samples were examined by acid-fast bacillus (AFB) smear microscopy using Ziehl–Neelsen staining on light microscope and graded according to the RNTCP guidelines19. Thereafter, the sputum samples were processed by modified Petroff's method19 and cultured on LJ medium. The slants were incubated at 37°C and were regularly observed for growth every week.
Biochemical confirmation of Mycobacterium tuberculosis: The growth obtained on L-J slants was identified by naked eye colony appearance as well as growth characteristics and was biochemically confirmed by niacin, nitrate test as well as susceptibility to p-nitrobenzoic acid20. Biochemically confirmed MTB isolates were subjected to DST testing as well as RAPD and MIRU VNTR typing.
Drug susceptibility testing: DST was performed by indirect one per cent proportion method on LJ media for the four first-line drugs (Sigma, USA), namely, streptomycin (4 μg/ml), isoniazid (INH) (0.2 μg/ml), rifampicin (40 μg/ml) and ethambutol (2 μg/ml)19,21.
Genomic DNA preparation: DNA was extracted from the isolates grown on LJ medium, by physiochemical method22. Briefly, the growth was scraped and suspended in 400 μl TE buffer [10 mM Tris-HCl (pH 8.0), 1 mM EDTA] and the above suspension was placed at 90°C for 20 min, to heat kill the bacteria. This was followed by snap chilling at -20°C for 20 min. Extracted DNA was precipitated overnight with isopropanol (-20°C) and washed with 70 per cent ethanol. The DNA was dissolved and stored in 40 μl TE buffer at 4°C till further usage. The purity and quantity of obtained DNA were checked on Nano-Quant (Tecan, Switzerland) for optimum quality. The quality checked DNA was diluted to a concentration of 25 ng/μl for RAPD analysis and 2 ng/μl for MIRU VNTR analysis.
Random amplified polymorphic DNA (RAPD) analysis: RAPD analysis was done by method described by Linton et al13. The DNA was amplified using single primer IS 986F - 5’ACG CTC AAC GCC AGA GAC CA 3’; Tm=61°C13 (Imperial Life Sciences, Gurgaon, India). This was targeted to the inverted repeat sequences of IS 986 between closely spaced copies of this element13. The PCR mix (25 μl) used for the experiment contained 1× PCR buffer, 1.5 mM MgCl2, 1.5 U of Taq DNA polymerase and 200 μM of deoxynucleoside triphosphate (dNTP). All reagents for PCR amplification were obtained from Merck, USA. To this mix, 50 ng of genomic DNA was added. The resulting mix was incubated for five minutes at 94°C. A total of 40 cycles of PCR was run on a thermal cycler (ABI, USA). The reaction consisted of a denaturation step for 30 sec at 94°C, an annealing step for one minute at 36°C and an extension step for one minute at 72°C. The annealing temperature was kept low to allow less stringent annealing of primer along the template DNA. This was followed by a step of final extension for seven minutes at 72°C. The analysis was repeated on the same DNA in another independent run experiment, following the same protocol to check whether the results were reproducible or not. The reproducible profiles were used in the analysis.
Mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU VNTR) analysis: For MIRU VNTR, 12 sets of primers were used for the MIRU loci17,23 [2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39, 40 (Imperial Life Sciences)]. The PCR mix was prepared using the HotStarTaq DNA polymerase kit (Qiagen, Germany). The reaction mixture (20 μl) contained 1× PCR buffer, 1.5-3.0 mM MgCl2, 0.4 U of Taq polymerase, 200 μM of dNTP mix, 0.4 μM of primer. The reactions were carried on ABI thermal cycler starting with a denaturing step of 15 min at 95°C. After denaturation, the PCR was performed for 40 cycles of one minute at 94°C, followed by one minute at 59°C and 1.5 min at 72°C. The reaction was terminated by incubating for 10 min at 72°C.
The PCR products generated by both the steps were visualized by electrophoresis in a gel containing two per cent agarose (Sigma, USA). For size determination, 100 bp (base pair) DNA marker (G Bio, India) was used. The reproducibility of the profile was checked by repeating the experiment twice.
Positive control (PC) & negative control (NC): Negative control consisted of PCR mix with nuclease-free distilled water instead of genomic DNA and was run with each batch of amplification. DNA from MTB-H37Rv was used as positive control (PC).
Analysis of fingerprints & cluster analysis: Gels were photographed and band sizes were estimated using a gel documentation and analysis system (Bio-Rad, USA). For RAPD analysis, the size of the bands that differed by less than 15 bp was considered to be the same. The band pattern for each isolate was noted. Only distinct and prominent bands were used in assessing RAPD patterns. For the non-reproducible RAPD patterns, the common bands from two independent runs were taken into account for cluster analysis. Taking into consideration all such bands obtained, the presence or absence of a band was noted for each isolate. A data matrix was composed and the numerals 1 and 0 were built on the basis of presence (1) or absence (0) of a band observed. This binary code for each isolate was used for the determination of phylogenetic similarity between isolates. The genetic relationship among the isolates was determined by constructing a similarity matrix based on calculating the Jaccard's coefficient11. Isolates with a Jaccard's coefficient of ≥0.8 were assumed to be clustered. Statistical analysis was performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA).
For MIRU VNTRs, the band sizes for the amplification products of each locus were determined. The number of repeats/alleles at each locus was determined using the MIRU VNTR allelic table17. This led to the formation of a 12-digit code for each isolate comprising the number of repeats at each locus. Two isolates with similar 12-digit code and /or difference at a single locus of the 12-digit number (i.e. >90% similarity) were grouped into one cluster. MIRU VNTR allelic diversity at each locus was determined using online tool (MIRU VNTR plus)23.
Results
Smear & culture positivity: Of the 175 sputum samples collected from patients under treatment, their contacts as well as suspected TB patients, culture results were obtained for 171 samples (four were contaminated and could not be retrieved). Among the 171 samples, 68 were from known pulmonary TB cases and 103 were from contacts which included family, neighbourhood, workplace and social contacts from the same slum area. Of these samples, 160 were untreated/ATT less than two months (NSP/CAT I patients) and 11 were retreatment cases (RT, CAT II patients). These comprised 106 males and 65 females (male:female=1.6:1), and their age ranged from 3 to 71 years (mean ±SD 35.46 ±16.91yr). All were HIV negative. On sputum smear microscopy, 75 of 171 (43.9%) were AFB positive. Mycobacterial growth on LJ culture was observed in 83 (48.5%) patients. Among the 103 suspected TB contacts, 17 (16.5%) were positive for MTB culture. All 83 isolates were biochemically confirmed as MTB. Seventy four of these patients (89.2%) were new untreated cases (CAT I) and nine (10.8%) were RT (CAT II). These CAT II cases were on treatment for varying duration ranging from six months to two years (Table I).
Table I.
Summary of bacteriological results of patients included in the study

Drug susceptibility testing (DST) patterns and treatment outcomes of patients: DST was done for all the 83 isolates. Among the 74 CAT I isolates, 66.2 per cent (n=49) were fully sensitive, three were MDR (4.1%); one (1.4%) were isoniazid monoresistant; nine (12%) were monoresistant to streptomycin and 12 (16%) were poly-drug resistant. Of the nine isolates from CATII cases, three (33.3%) were fully sensitive; two (22%) were MDR; three (33%) were poly-drug resistant and one (11%) was mono-resistant to isoniazid (Table II).
Table II.
Drug susceptibility profiles of culture-positive cases
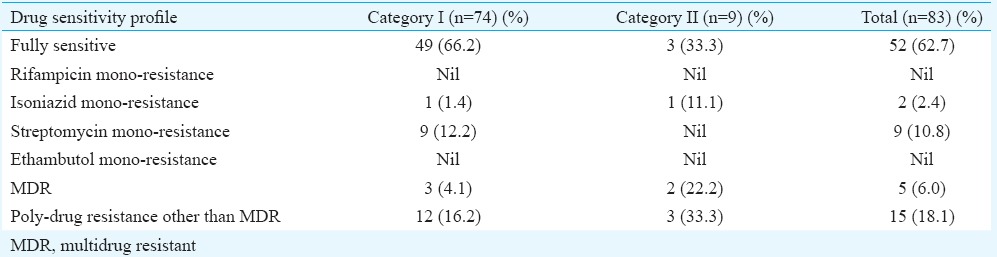
Mono-resistance to rifampicin and ethambutol was not seen in both the categories of patients. Mono-resistance to streptomycin was quite high (12.2%) in CAT I patients but was not observed in CAT II patients. Poly-drug resistance was observed in 16.2 and 33.3 per cent, respectively, in CAT I and CAT II patients (Table II).
Random amplified polymorphic DNA (RAPD): RAPD analysis and MIRU VNTR fingerprints could be typed for 81 of 83 isolates (97.6%). RAPD profiles of the isolates showed band patterns consisting of 1-15 bands, detected in the range of 176-1600 bp. Positive control, MTB-H37Rv showed two prominent bands at 810 bp and at 377 bp in all amplifications (Fig. 1). A total of 64 patterns were obtained (58 unique and 6 clustered). Twenty three isolates were clustered in these six clusters (28.4%), whereas 58 (71.6%) isolates showed unique pattern. The largest cluster was of seven isolates (Table III).
Fig. 1.
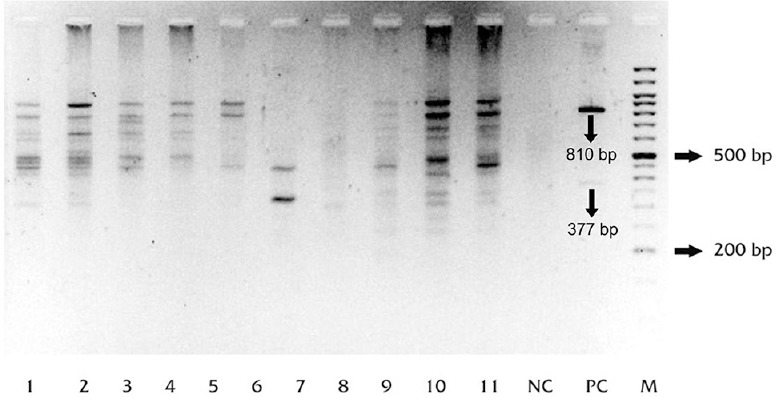
Agarose gel image showing random amplified polymorphic DNA (RAPD) profiles of Mycobacterium tuberculosis (MTB) isolates from the two slums. Lanes 1-11, samples (isolates from the slums); NC, no-template control; PC, positive control (M. tuberculosis strain H37Rv).
Table III.
Epidemiological links & drug susceptibility testing (DST) patterns in random amplified polymorphic DNA (RAPD) clusters
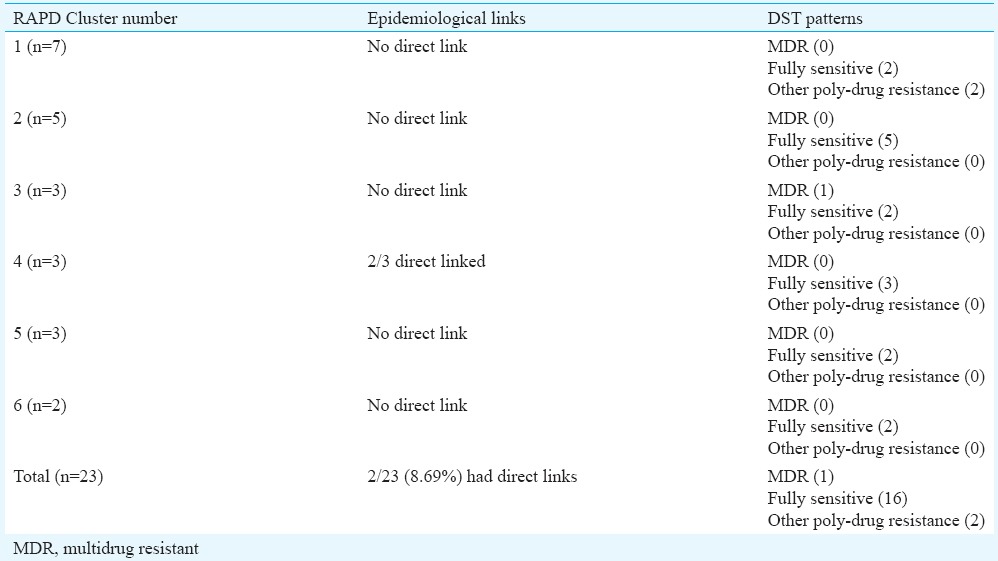
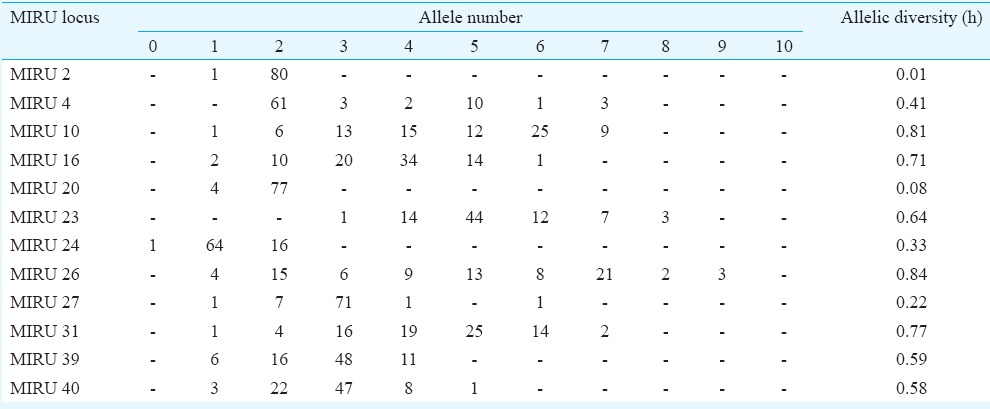
Mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU VNTR) analysis: MIRU VNTR analysis revealed 69 patterns (61 unique & 8 clustered). Lineage of 52 of 81 (64.19%) isolates could not be determined by this typing. No association was observed with VNTR and Beijing or any other lineage in the MDR isolates (Table IV). MIRU VNTR signature code of H37Rv, i.e. 23’3226133321, was observed in all amplifications (Fig. 2). In all, 20 (24.7%) isolates were clustered and 61 were unique (75.3%). The Hunter Gaston's Discriminatory index (HGDI)23 for RAPD was 0.9873 and for MIRU VNTR was 0.9951. Five MIRU loci (10, 16, 23, 26, 31) were found to be ‘highly discriminatory’ (allelic diversity; h≥0.6), four MIRU loci (4, 24, 39, 40) were ‘moderately discriminatory’ (h: 0.3-0.59) and three loci (2, 20, 27) were ‘poorly discriminatory’ (h< 0.3) (Table V). The concordance between MIRU VNTR and RAPD clusters was low. In all, 14 (17.3%) isolates were clustered by RAPD alone, 11 (13.6%) by MIRU VNTR alone and nine (11.1%) were clustered by both the methods whereas 47 (58.02%) were not clustered by either of the methods. Among the 23 isolates obtained from known cases and epidemiologically linked contacts, only two (8.9%) were clustered by RAPD and nine (39.1%) by MIRU VNTR.
Table IV.
Epidemiological links & drug susceptibility testing (DST) patterns in mycobacterial interspersed repetitive unit-variable number of tandem repeat (MIRU VNTR) clusters
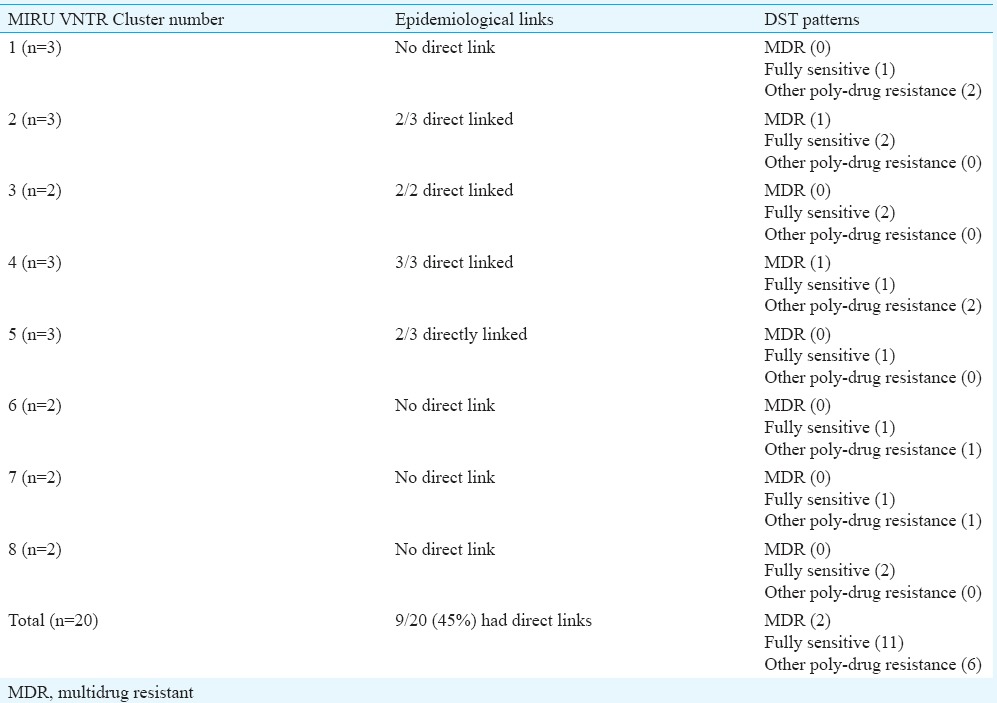
Fig. 2.
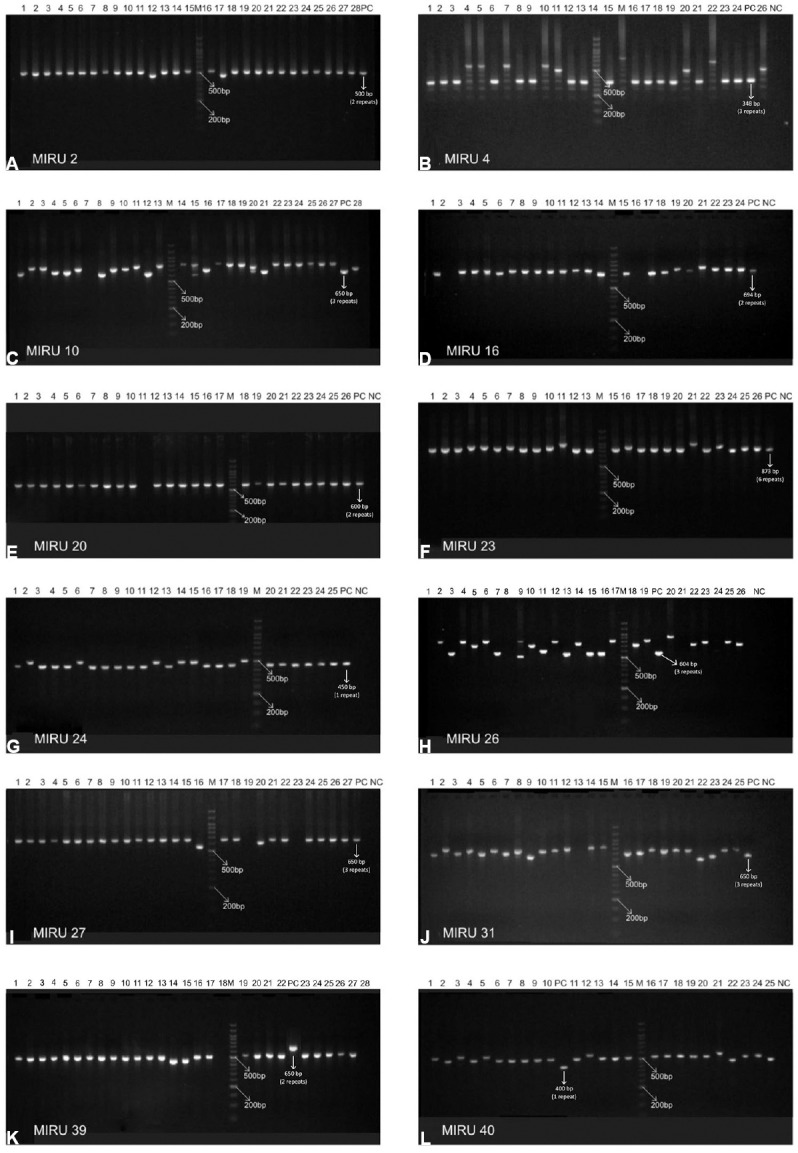
Agarose gel images of mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU VNTR). Panels (A-L) depict MIRU 2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39 and 40. In each panel lanes are marked as M, 100 bp DNA marker; various lanes, MIRU VNTR profiles of the isolates; NC, MIRU fingerprint of no-template control for the respective locus; PC, MIRU fingerprint of positive control (M. tuberculosis strain H37Rv) for the respective locus.
Table V.
Mycobacterial interspersed repetitive unit-variable number of tandem repeat (MIRU-VNTR) allelic diversity
Discussion
In the present study, 48.5 per cent samples collected from known pulmonary TB patients, suspects and their contacts were positive for MTB. The AFB smear and culture positive rates were low as compared to that reported in the general population by RNTCP under the programme conditions1. This could be due to inclusion of a section of patients who were also on treatment in the slums but had taken less than two months treatment. Similar culture positive rates have been reported from other parts of the country and in other slum settings from Lucknow24, Gujarat25 and south India26.
About 66.2 per cent of isolates belonging to NSP cases (CAT I) were fully sensitive to all of the four drugs tested in the present study. Studies undertaken at Lucknow24, Gujarat7,25 and Ernakulam (Kerala)26 have reported higher sensitivity to all the first-line drugs (78.6, 78.7 & 72.1%, respectively). MDR TB observed in the present study in the NSP cases was 4.1 per cent, which was comparable to those from Lucknow24 (4.7%) but higher than Gujarat25 (2.4%), and Kerala26 (0.6%). Poly-drug resistance other than MDR was seen in 16.2 per cent of our isolates, which was higher than other studies which reported 3 to 10.7 per cent9,24,25,26. Total resistance (both mono- and poly-drug) to streptomycin in CAT I cases was high, i.e. 33.63 per cent, as also reported by other workers from India (27.69 & 70.0%8, respectively). This could be due to indiscriminate use of streptomycin in slums which is a cheap injectable as well as readily available drug from the pharmacist.
Acquired multidrug resistance in the present study was observed to be 22.2 per cent; poly-drug resistance other than MDR 33.3 per cent and mono-drug resistance to INH as 11.1 per cent. The profile may not be truly representative as very few sputum samples from CATII patients were analysed. Study from Gujarat25 reported 17.4 per cent MDR, other poly-drug resistance eight per cent and mono-drug resistance to INH as 11.1 per cent.
Isolates with MDR pattern in the present study were similar to our previous report, i.e. 4.5 and 24.3 per cent27 in NSP and RT cases, respectively, and marginally higher (2.5 & 16%, respectively)1 than the reported national average. Mono-resistance to streptomycin was high (12.2%) in CAT I patients but was not observed in CAT II patients. As the study had only a small number of CAT II cases, no conclusions could be drawn. The high percentage of poly-drug resistance observed in both CATI and CATII patients needs to be addressed, as these patients are more commonly treatment failure cases and are prone to develop into MDR.
PCR-based genotyping methods are easy to use, rapid and need one day. The RFLP-based methods require 3-4 days and more labour intensive. RAPD analysis uses single randomly selected primer to scan the entire bacterial genome, therefore can be applied to any genome irrespective of its complexity and has been used by many Indian investigators4,28,29,30 for DNA fingerprinting. Using RAPD analysis, a previous study from Agra clustered 9 of 16 (56.25%) isolates obtained from the same family residing in a slum area29.
The reliability and reproducibility of the method reside in the correct choice of primers and optimization of reaction conditions. IS 986 F primer is targeted to the inverted repeat sequences of the insertion sequence 98613. In our study, the fingerprints obtained from two independent runs on the same DNA sample yielded reproducible results in only 55 of 81 (68%) isolates. Differences between the patterns generated by RAPD might be due to technical and operating parameters of the method rather than true inter genetic polymorphism. Variations in RAPD patterns are attributed to priming efficiency during early rounds of amplification, concentration and purity of template DNA, primer / template ratio, or the ramp times of the cyclers used31,32,33. In many settings like slums, tuberculosis often results from casual contact, in addition to close contact with infected patients13. Standard MIRU VNTR typing has been shown to have slightly good predictive value for tracing TB transmission32, and clustering has shown epidemiological linkages and has higher reproducibility and discriminatory power18. In our study also, discriminatory power of MIRU VNTR analysis was higher (HGDI=0.9951) in comparison with RAPD analysis (HGDI=0.9873). Studies from Kanpur18, New Delhi34 and Mumbai35 also reported that MIRU 26 was highly discriminatory using the 12 loci. Although the 24 locus variant of the method is more discriminatory, the present study being the first one from this region reports the use of 12 loci. Moreover, 24 loci MIRU VNTR has been found to be more useful for phylogenetic studies whereas 12 and 15 loci MIRU VNTR has been found useful for epidemiological investigations33. In the present study, 28.39 per cent isolates were clustered by RAPD analysis and 24.69 per cent by MIRU VNTR analysis. However, among the known contacts, only 8.9 per cent were clustered together by RAPD while 39.1 per cent by MIRU VNTR.
No association was observed in genotypic clustering and drug resistance patterns. Poly-drug resistance was observed in only one cluster by RAPD, but by MIRU, 50 per cent clusters showed poly-drug resistance. Higher clustering of MTB isolates from slums has been reported by several workers2,3. The lower rates of clustering (25-28.0%) found in our study may be because of inability to include all TB patients in the areas as well as that the infection may have been acquired from outside the slums.
Limitations of our study were the inability to include all TB patients, high mobility of inhabitants in the slum areas leading to possible missing of clustered cases, reactivation of old dormant cases exposed years ago and thus failure to establish links. The benefit of the study was that it tried to analyse the drug resistance profile in new and retreated TB cases and genetic diversity in slum areas. MDR rates observed were similar to the rest of country.
In conclusion, rifampicin and ethambutol mono-resistance was not seen in MTB isolates and only six per cent isolates were MDR. MIRU VNTR technique was found to be highly discriminatory and efficient in identifying definitive epidemiologically linked cases. RAPD analysis was rapid, inexpensive, had fairly high discriminatory power but had lower reproducibility and was unable to link related patients in the community.
Acknowledgment
Training and technical support provided by the National JALMA Institute for Leprosy & Other Mycobacterial Diseases, Agra, and financial support from Indian Council of Medical Research, New Delhi (5/8/5/1/2008 ECD-1 DT 18/5/09) are gratefully acknowledged.
Footnotes
Conflicts of Interest: None.
References
- 1.WHO (2016) Global tuberculosis report. Geneva Switzerland: World Health Organization; 2016. [accessed on November 17, 2016]. Available from: http://apps.who.int/iris/bitstream/10665/250441/1/9789241565394-eng.pdf . [Google Scholar]
- 2.Mendes JM, Machado SMA, Lourenço MC, Ferreira RMC, Fonseca Lde S, Saad MHF. Molecular diversity of Mycobacterium tuberculosis strains in a slum area of Rio de Janeiro, Brazil. J Bras Pneumol. 2008;34:1063–8. doi: 10.1590/s1806-37132008001200012. [DOI] [PubMed] [Google Scholar]
- 3.Dowdy DW, Golub JE, Chaisson RE, Saraceni V. Heterogeneity in tuberculosis transmission and the role of geographic hotspots in propagating epidemics. Proc Natl Acad Sci U S A. 2012;109:9557–62. doi: 10.1073/pnas.1203517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain A, Mondal R, Prasad R, Singh K, Ahuja RC. Prevalence of multidrug resistant Mycobacterium tuberculosis in Lucknow, Uttar Pradesh. Indian J Med Res. 2008;128:300–6. [PubMed] [Google Scholar]
- 5.Tyagi K, PRIA, SPARC. Status and Opportunities of Development for Urban Poor/Slums Dwellers in Jaipur City. 2012. [accessed on November 11, 2013]. Available from: http://apps.who.int/iris/bitstream/10665/250441/1/9789241565394-eng.pdf .
- 6.Kumar M. Need assessment for urban health in slums of Jaipur. Indian J Res Econ Soc Sci. 2013;3:52–63. [Google Scholar]
- 7.Kadri AM, Bhagyalaxmi A, Lala MK, Jivrajini P, Vidhani M, Patel T. An epidemiological study of prevalence of tuberculosis in the urban slum area of Ahmedabad city. Indian J Community Med. 2003;28:122–4. [Google Scholar]
- 8.Nair DM, George A, Chacko KT. Tuberculosis in Bombay: New insights from poor urban patients. Health Policy Plan. 1997;12:77–85. doi: 10.1093/heapol/12.1.77. [DOI] [PubMed] [Google Scholar]
- 9.Singh V, Jaiswal A, Porter JDH, Ogden JA, Sarin R, Sharma PP, et al. TB control, poverty, and vulnerability in Delhi, India. Trop Med Int Health. 2002;7:693–700. doi: 10.1046/j.1365-3156.2002.00909.x. [DOI] [PubMed] [Google Scholar]
- 10.Banu S, Rahman MT, Uddin MKM, Khatun R, Ahmed T, Rahman MM, et al. Epidemiology of tuberculosis in an urban slum of Dhaka City, Bangladesh. PLoS One. 2013;8:e77721. doi: 10.1371/journal.pone.0077721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. Molecular epidemiology of tuberculosis: Current insights. Clin Microbiol Rev. 2006;19:658–85. doi: 10.1128/CMR.00061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Deutekom H, Supply P, de Haas PE, Willery E, Hoijng SP, Locht C, et al. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J Clin Microbiol. 2005;43:4473–9. doi: 10.1128/JCM.43.9.4473-4479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linton CJ, Jalal H, Leeming JP, Millar MR. Rapid discrimination of Mycobacterium tuberculosis strains by random amplified polymorphic DNA analysis. J Clin Microbiol. 1994;32:2169–74. doi: 10.1128/jcm.32.9.2169-2174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chauhan DS, Singh D, Sharma VD, Das R, Singh HB, Srivastava K, et al. Molecular typing of Mycobacterium tuberculosis isolates from different parts of India based on IS 6110 element polymorphism using RFLP analysis. Indian J Med Res. 2007;125:577–81. [PubMed] [Google Scholar]
- 15.Das S, Paramsivan CN, Lowrie DB, Prabhakar R, Narayanan PR. IS 6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, south India. Tuber Lung Dis. 1995;76:550–4. doi: 10.1016/0962-8479(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan I, Manju VK, Kumar AR, Mundayoor S. Implications of low frequency of IS 6110 in fingerprinting field isolates of M. tuberculosis from Kerala, India. J Clin Microbiol. 2001;39:1683. doi: 10.1128/JCM.39.4.1683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Supply P. Multilocus Variable Number Tandem Repeat Genotyping of Mycobacterium tuberculosis. Technical Guide. Institut de Biologie/Institut Pasteur de Lille. 2005. [accessed on March 27, 2012]. Available from: http://www.miru-vntrplus.org/MIRU/files/MIRU-VNTRtypingmanualv6.pdf .
- 18.Sharma P, Chauhan DS, Upadhyay P, Faujdar J, Lavania M, Sachan S, et al. Molecular typing of Mycobacterium tuberculosis isolates from a rural area of Kanpur by spoligotyping and mycobacterial interspersed repetitive units (MIRUs) typing. Infect Genet Evol. 2008;8:621–6. doi: 10.1016/j.meegid.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 19.RNTCP. Training Manual for Mycobacterium tuberculosis Culture & Drug susceptibility testing. 2009. [accessed on July 15, 2011]. Available from: http://www.tbcindia.nic.in/pdfs/Training%20manual%20M%20tuberculosis%20C%20DST.pdf .
- 20.Kent PT, Kubica GP. Public health mycobacteriology: A guide for the level III laboratory. Atlanta, GA: US Health Service Centers for Disease Control; 1985. [Google Scholar]
- 21.Canetti G, Fox W, Khomenko AA, Mahler HT, Menon NK, Mitchison DA, et al. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ. 1969;41:21–43. [PMC free article] [PubMed] [Google Scholar]
- 22.van Soolingen D, de Haas PE, Hermans PW, van Embden JD. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1994;235:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- 23.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–9. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta H, Kant S, Jain A, Natu SM, Ahluwalia S. Initial drug resistance pattern among pulmonary tuberculosis patients. Indian J Tuberc. 2013;60:154–61. [PubMed] [Google Scholar]
- 25.Ramachandran R, Nalini S, Chandrasekar V, Dave PV, Sanghvi AS, Wares F, et al. Surveillance of drug-resistant tuberculosis in the state of Gujarat, India. Int J Tuberc Lung Dis. 2009;13:1154–60. [PubMed] [Google Scholar]
- 26.Joseph MR, Shoby CT, Amma GR, Chauhan LS, Paramasivan CN. Surveillance of anti-tuberculosis drug resistance in Ernakulam District, Kerala State, South India. Int J Tuberc Lung Dis. 2007;11:443–9. [PubMed] [Google Scholar]
- 27.Malhotra B, Pathak S, Vyas L, Katoch VM, Srivastava K, Chauhan DS, et al. Drug susceptibility profiles of Mycobacterium tuberculosis isolates at Jaipur. Indian J Med Microbiol. 2002;20:76–8. [PubMed] [Google Scholar]
- 28.Singh HB, Chauhan DS, Singh D, Das R, Srivastava K, Yadav VS, et al. Rapid discrimination of Indian isolates of M. tuberculosis by random amplified polymorphic DNA (RAPD) analysis – A preliminary report. Indian J Med Microbiol. 2002;20:69–71. [PubMed] [Google Scholar]
- 29.Singh HB, Chauhan DS, Faujdar J, Upadhyay P, Saxena N, Yadav VS, et al. Tracing transmission of tuberculosis by random amplified polymorphic DNA (RAPD) analysis within same family & neighbourhood. Indian J Med Res. 2007;126:82–4. [PubMed] [Google Scholar]
- 30.Singh Y, Katoch VM. Analysis of INH drug resistance among Mycobacterium tuberculosis strains using RAPD-PCR. Indian J Comp Microbiol Immunol Infect Dis. 2008;29:27–30. [Google Scholar]
- 31.Roetzer A, Schuback S, Diel R, Gasau F, Ubben T, di Nauta A, et al. Evaluation of Mycobacterium tuberculosis typing methods in a 4-year study in Schleswig-Holstein, Northern Germany. J Clin Microbiol. 2011;49:4173–8. doi: 10.1128/JCM.05293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwara A, Schiro R, Cowan LS, Hyslop NE, Wiser MF, Harrison SR, et al. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003;41:2683–5. doi: 10.1128/JCM.41.6.2683-2685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagielski T, van Ingen J, Rastogi N, Dziadek J, Mazur PK, Bielecki J. Current methods in the molecular typing of Mycobacterium tuberculosis and other mycobacteria. Biomed Res Int. 2014;2014:645802. doi: 10.1155/2014/645802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stavrum R, Myneedu VP, Arora VK, Ahmed N, Grewal HM. In-depth molecular characterization of Mycobacterium tuberculosis from New Delhi – Predominance of drug resistant isolates of the ‘modern’ (TbD1) type. PLoS One. 2009;4:e4540. doi: 10.1371/journal.pone.0004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterjee A, Mistry N. MIRU-VNTR profiles of three major Mycobacterium tuberculosis spoligotypes found in Western India. Tuberculosis (Edinb) 2013;93:250–6. doi: 10.1016/j.tube.2012.10.004. [DOI] [PubMed] [Google Scholar]


