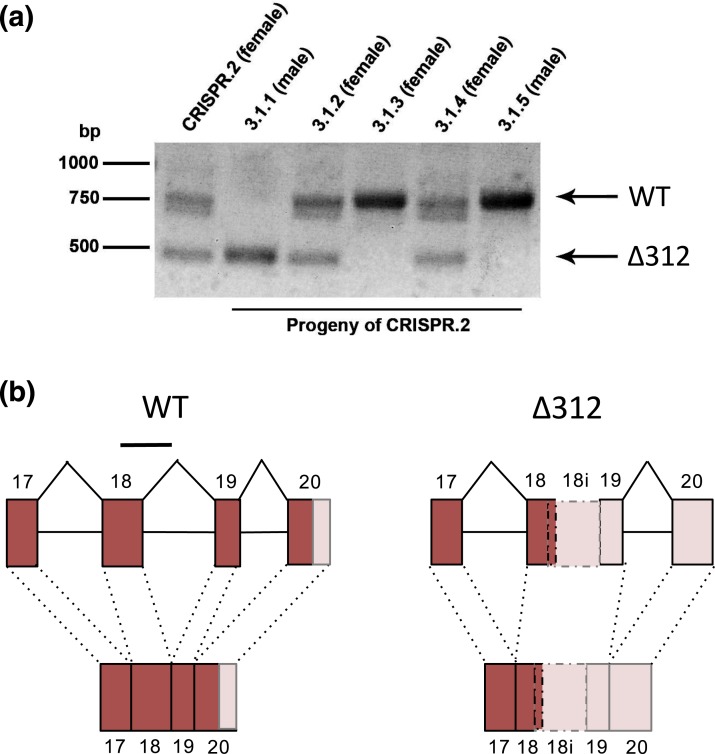
Figure 1.
Generation of Igsf1 loss-of-function mice using CRISPR-Cas9. (a) PCR amplification of the targeted region in exon 18 of Igsf1 in the founder female (CRISPR.2, lane 1) and five of her progeny (3.1.1 to 3.1.5, lanes 2 to 6). Arrows at the right indicate the wild-type (top) or Δ312 (bottom) alleles. (b) Genomic organization (top) and RNA splicing (bottom) around exons 17 to 20 of the wild-type (left) and Δ312 alleles (right). Exons are presented as boxes and are numbered. Intervening introns are shown as lines. The deleted parts of exon 18 and intron 18 in the Δ312 allele are shown with a line at the top of the wild-type schematic (left). The novel exon in the Δ312 allele, which is derived from the 5′ end of exon 18, the 3′ end of intron 18 (labeled 18i), and all of exon 19, is shown with broken lines (right). Dark boxes denote translated sequences, whereas the pale boxes reflect untranslated regions. Genomic structure 5′ of exon 17 is not pictured. WT, wild-type.