Figure 8.
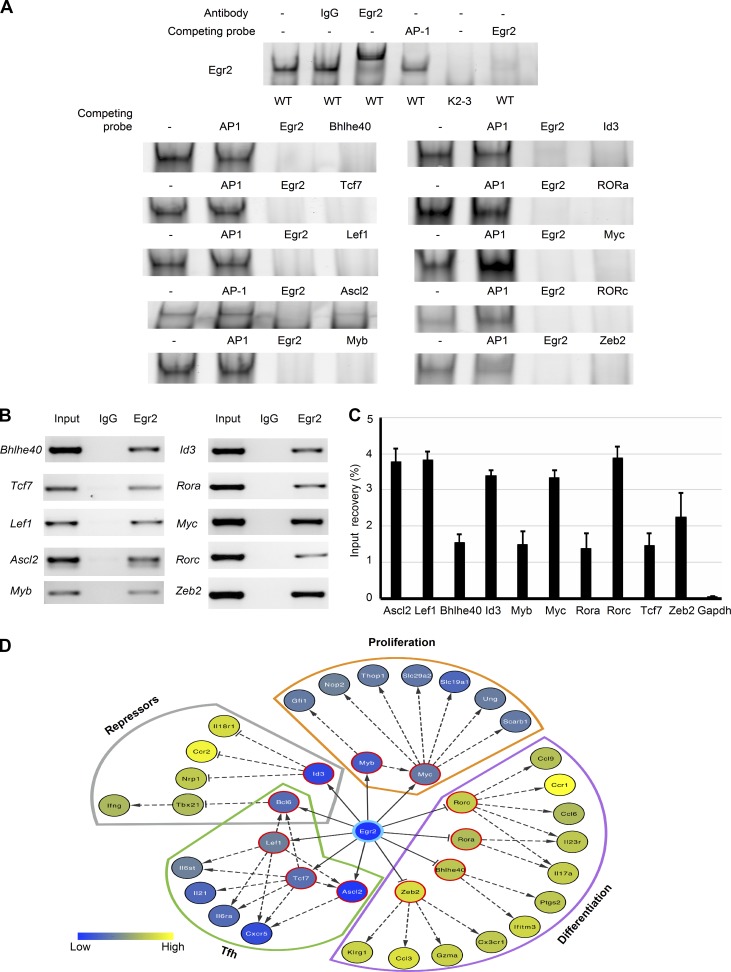
Egr2 binds to and regulates the expression of transcription factors involved in the proliferation and differentiation of T cells. (A) CD4 T cells from WT mice were stimulated for 16 h in vitro with anti-CD3 and anti-CD28 and nuclear proteins used for EMSA analysis. Probes derived from the potential Egr2-binding sites in the loci of the indicated transcription factors were used as competitors for the interaction of Egr2 with a labeled probe containing its consensus binding sequence. Unlabeled probe containing the AP-1 consensus sequence served as negative control, and unlabeled Egr2 consensus sequence served as positive control. The specificity of the band was confirmed by supershift. Nuclear proteins from stimulated CD2-Egr2/3−/− (K2-3) CD4 T cells served as a further negative control. (B and C) ChIP assay of anti-Egr2 precipitates from anti-CD3 and anti-CD28 stimulated CD4 T cells from WT mice. Total chromatin extracts from the same cells were used as positive controls, and IgG precipitates served as negative controls. ChIP data are presented as the percentage recovery of input in C. (D) Model of the “Egr2-centric” gene expression network in T cells in response to viral infection. Nodes are colored according to the level of expression in K2-3 cells in the RNA-seq data (Fig. 6), and those with a red border are direct Egr2 target genes. A–C are representative of three independent experiments. Data in C are means ± SD.