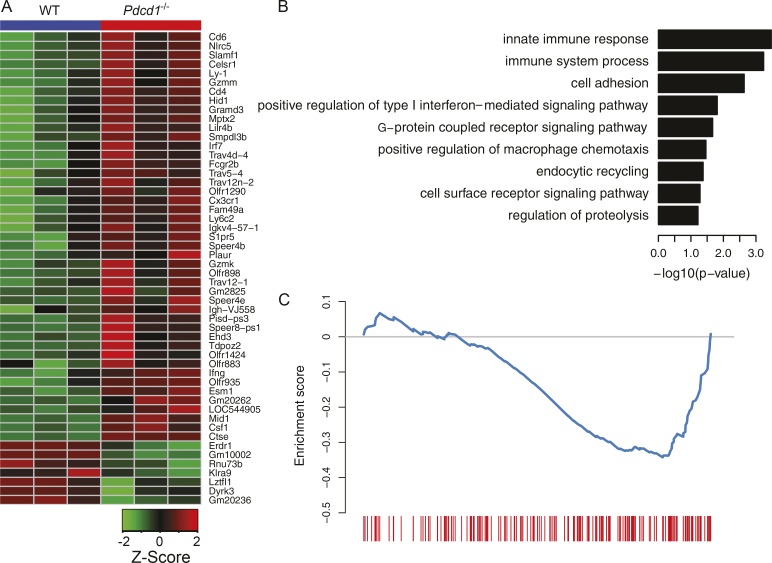
Figure 4.
Comparison of gene expression profiles of KLRG1+ ILC-2s between WT and Pdcd1−/−. Lin−CD90+KLRG1+ ILC-2s were sorted from the lungs of WT or Pdcd1−/− mice that had been treated with IL-33 for 3 d. Total RNA was isolated, and then subjected to microarray analysis. Heat map for the expression patterns of differentially expressed genes (A). Representative GO terms enriched for genes up-regulated in Pdcd1−/−. Only GO terms from the biological process category is shown. The log10-scaled p-value is indicated in the x axis (B). Gene set enrichment analysis of the effect of Pdcd1 knockout for gene expression. The genes are ranked by fold change as evaluated by microarray data, and compared with the gene set available from GEO under accession no. GSE26495 (NAIVE_VS_PD1LOW_CD8_TCELL_DN). The blue curve shows the running enrichment score (ES) for the gene set as the analysis walks down the ranked gene list. The leading edge for the gene set is shown as short bars at the bottom. The normalized enrichment score (NES) is calculated as –1.51, and the false discovery rate FDR is calculated as 0.036 (C). A one-way ANOVA analysis was performed to determine statistical significance. Microarray analysis was performed in triplicates, and each experiment consisted of n = 5 mice per cohort.