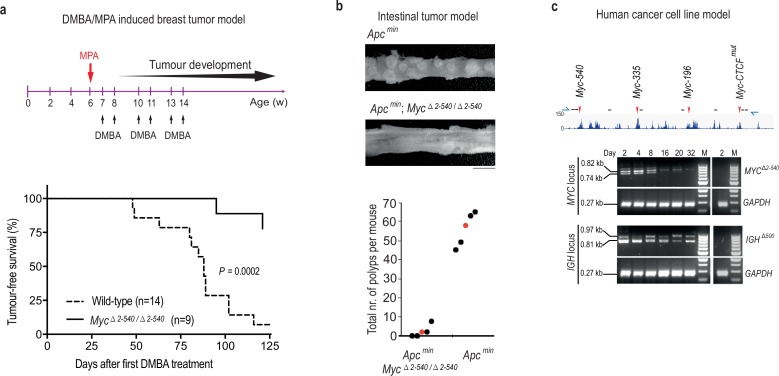
Figure 6. Myc −2 to −540 kb genomic region is required for the growth of cancers in vivo and cancer cells in vitro.
(a) Tumor-free survival plots showing resistance of Myc△2–540/△2–540 mice to development of DMBA/MPA induced mammary tumors. p-value=0.0002 (Mantel-Cox Log-rank test). See Figure 6—source data 1 for details. (b) The Myc −2 to −540 kb deletion results in fewer polyps than the Myc-335 deletion alone. p-value=0.00019 (Students T-test, 2-tailed). Apcmin mice were of 4 months of age (n = 5) and Apcmin; Myc△2–540/△2–540 mice were 6 months old (n = 5) at the time of analysis. Filled circles correspond to individual mice and red color denotes the median. See Figure 6—source data 1 for details. Bar equals 5 mm. (c) Crispr-Cas9 mediated deletion of region corresponding to Myc△2–540/△2–540 in human GP5d colon cancer cells, results in a loss of the edited cells over time. Top panel shows the active enhancer elements in GP5d cells within this region as determined by ChIP-seq analysis of histone H3 lysine 27 acetylation (H3K27ac). The sites of sgRNAs (black lines) and genotyping primers (blue arrows) used are indicated (not to scale). Red arrows mark the enhancer regions used in this study. Bottom panel shows the PCR-genotyping of the MYC locus and the control IGH locus showing the specific loss of the cells with the edited MYC locus over time. GAPDH was used as internal control. The right panel in each set shows absence of any deletion in the non-transfected cells (day 2). 100 bp ladder DNA molecular weight marker is shown (M).
DOI: http://dx.doi.org/10.7554/eLife.23382.015