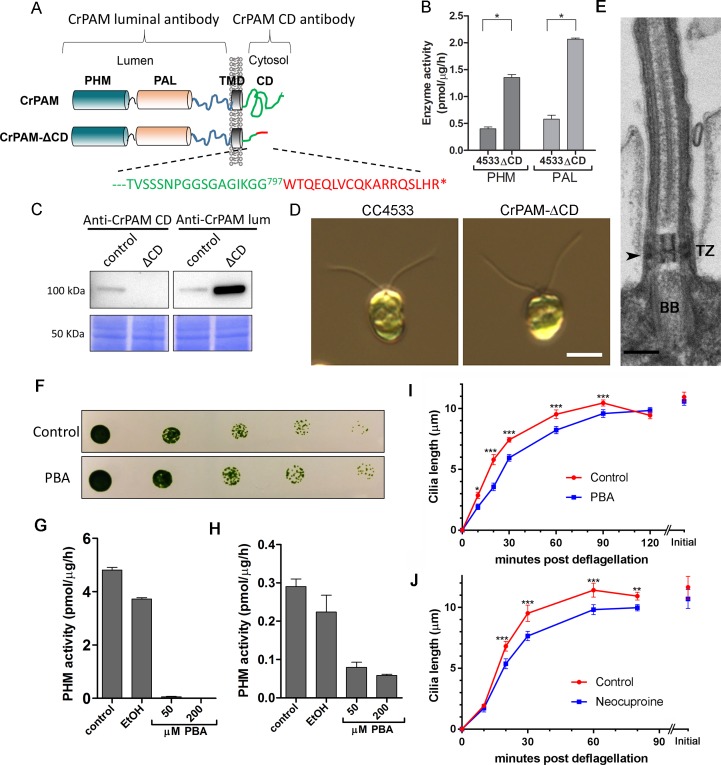
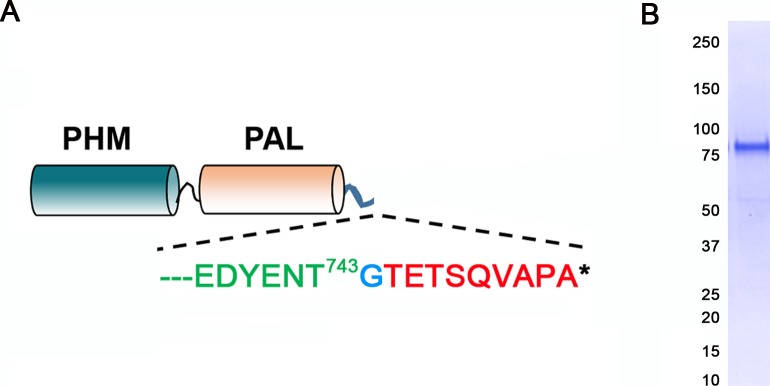
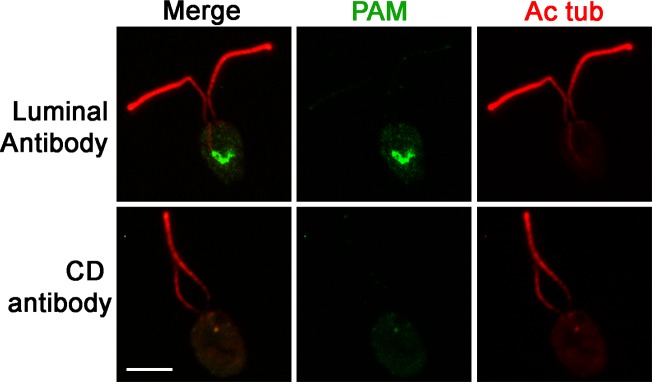
Figure 2. C. reinhardtii strain lacking the C-terminal domain of PAM assembles cilia.
(A) Schematic showing disruption of the cytosolic domain of CrPAM in the CrPAM-ΔCD strain. Residues corresponding to the C-terminus of CrPAM (green) and insertion cassette (red) are shown. (B) PHM and PAL specific activities in control (CC-4533) and CrPAM-ΔCD strain lysates (mean ± SD). Both PHM and PAL activity increased in CrPAM-ΔCD cells (p<0.05 using unpaired t-test). (C) Western blot of control and CrPAM-ΔCD strain lysates with C-terminal domain and luminal antibodies. As predicted by its sequence, CrPAM-ΔCD could be detected by the PAM luminal domain antibody, but not by the PAM cytosolic domain antibody. (D) Differential interference contrast images of control and CrPAM-ΔCD cells showing the presence of cilia in both (Scale bar, 5 µm). (E) Transmission electron micrograph showing normal ciliary structure in CrPAM-ΔCD strain. Basal body (BB), transition zone (TZ) and Y-linkers (arrowhead) appear normal (Scale bar, 200 nm). (F) Growth of 5-fold serially diluted CC-124 wildtype C. reinhardtii on TAP plates containing 0 (control) or 200 μM phenylbutenoic acid (PBA) 6 days post-innoculation. (G and H) Cell lysates were prepared from mouse corticotropes expressing rat PAM (G) and from C. reinhardtii CC-124 cells (H); PHM assays were carried out after addition of the indicated amount of PBA or vehicle (ethanol, EtOH). PHM specific activity was greatly reduced in both lysates in the presence of PBA. (I and J) Wildtype C. reinhardtii CC-124 cells were preincubated for 3 hr with PBA or neocuproine, deciliated by pH shock, which releases cilia immediately distal to the transition zone, and allowed to regrow cilia in the presence of 200 μM PBA (I) or 10 μM neocuproine (J). Error bars represent 95% confidence intervals; 80–100 measurements were made for each point (***p<0.001; **p<0.01; *p<0.05). Differences between control and experimental plots are significant (unweighted means analysis; p<0.0001; variance = 1.94% for PBA and 2.07% for neocuproine). Graphs are from one of two experiments which yielded similar results.