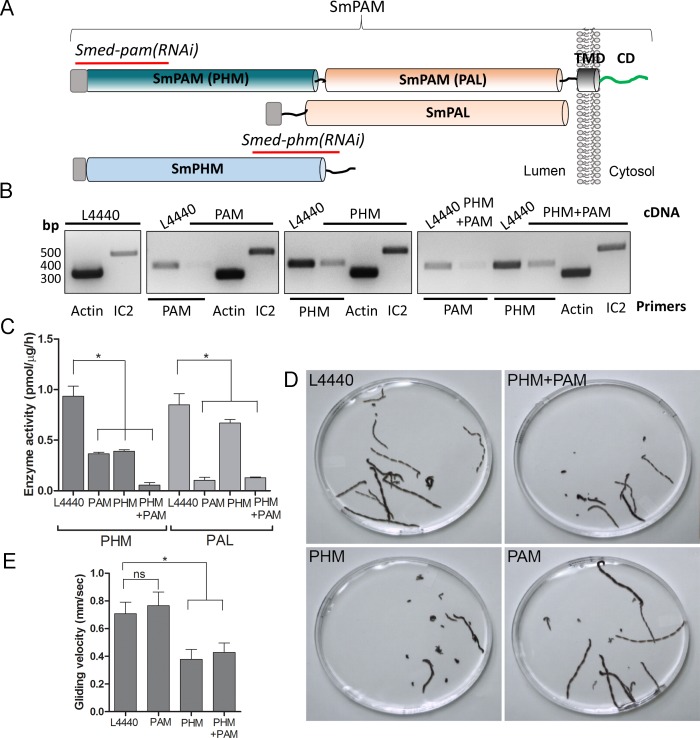
Figure 4. Knockdown of PHM gene in planaria causes motility defects.
(A) Schematic of S. mediterrania bifunctional membrane PAM, monofunctional PAL and monofunctional soluble PHM showing the regions used to generate RNAi constructs (red lines). Signal sequences, grey; transmembrane domain in SmPAM, black; cytosolic domain, CD, green. (B) RT-PCR analysis of mRNA from flatworms fed with the L4440 (control), PAM, PHM or PHM+PAM dsRNA. The cDNA source is shown above the image, and the primer pairs used for RT-PCR are listed below the image. Actin and dynein IC2 primers were used as controls. (C) PHM and PAL specific activities in planarian lysates plotted as mean ± SD from triplicate assays (asterisks indicate p<0.05 in a one-way Anova). (D) Overlays of frames from deconvolved videos of control and RNAi planaria showing the tracks taken over 60 s. (E) PHM and PHM+PAM RNAi animals displayed reduced gliding velocities plotted as mean ± SEM compared to L4440 and PAM RNAi animals (asterisks indicate p<0.05 in an unpaired t test).