FIG 4 .
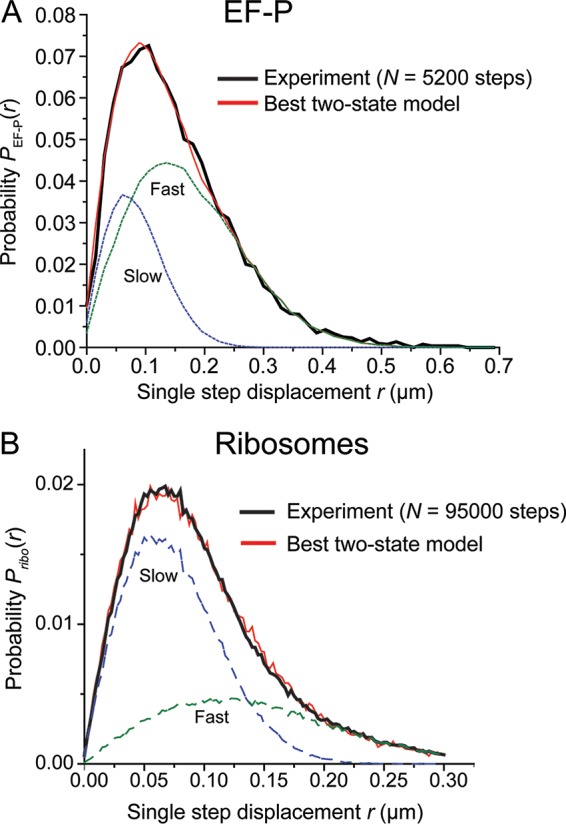
(A) Black, experimental probability distribution of single-step displacements taken by EF-P–mEos2 molecules in 2 ms. Red, the best fit to a static two-state model (without transitions) with Dslow constrained. Model parameters: Dslow = 0.2 µm2/s (σslow = 50 nm), fslow = 0.30, Dfast = 4.3 µm2/s (σfast = 75 nm), ffast = 0.7, with a χν2 = 1.0. The individual slow and fast components are shown in dashed lines as labeled. (B) Black, experimental probability distribution of single-step displacements taken by ribosomes (30S labeled with mEos2) in 2 ms. Red, the best unconstrained fit to a static two-state model (without transitions). Model parameters: Dslow = 0.20 µm2/s (σslow = 40 nm), fslow = 0.65, Dfast = 0.8 µm2/s (σfast = 75 nm), ffast = 0.35, with χν2 = 2.3. The slow and fast components are shown as dashed lines as labeled.
