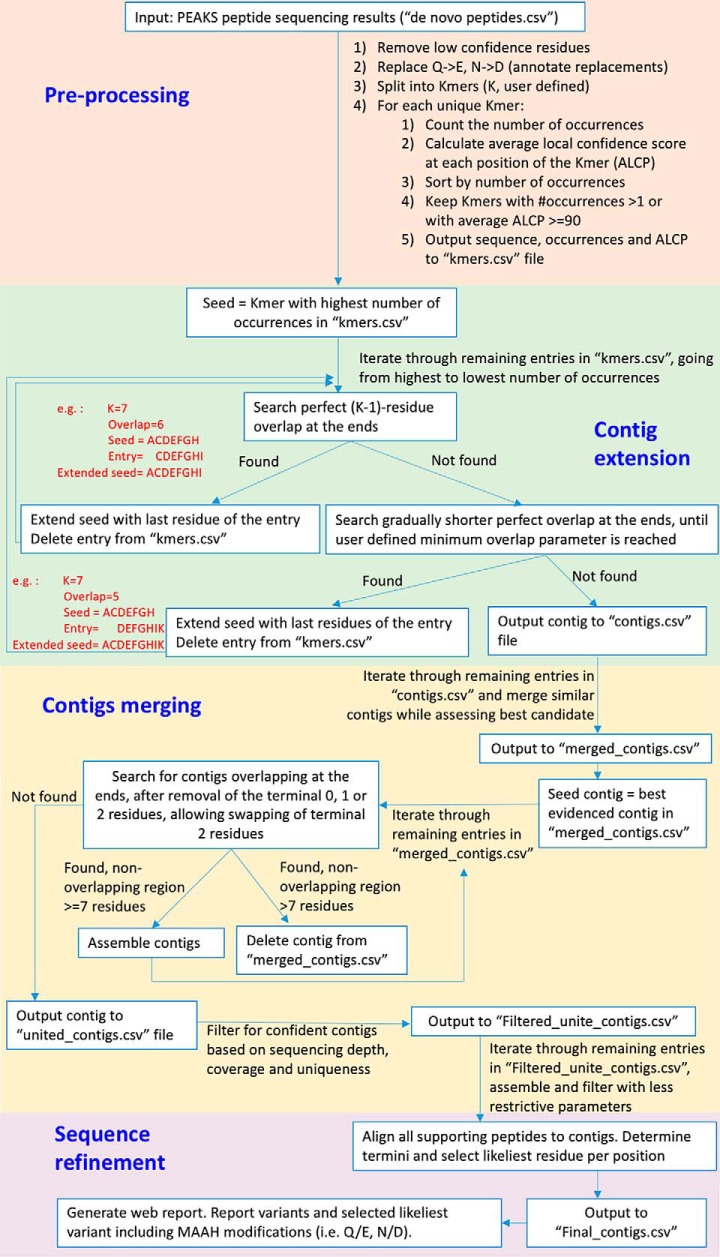
Fig. 2.
pTA algorithm design and rationale. The input to pTA is the unfiltered PEAKS de novo peptide sequencing output. We define a K-mer as a peptide with K amino acids. pTA has four stages: stage 1, pre-processing, aimed at maintaining a maximal number of peptide tags while removing low confident sequence assignments. Rationale, noise filtering reduces assembly of incorrect tags. Stage 2, contig extension, each K-mer that has not been assembled yet is used as a seed for assembling the longest possible contig, using all original K-mers. Rationale, occasionally the correct K-mer peptide will not have the highest number of occurrences and will not be used for assembly, unless used as a seed. Stage 3, contig merging, contigs are aligned and merged to produce the longest possible sequence. Rationale, premature termination of contig assembly (pTA stage 2) is due to extension of the growing contig by incorrect residues. The correct sequence, overlapping the termination point, can be found in another contig seeded by the correct K-mer or extended in the opposite direction (assembly of the correct sequence will not terminate at that point because there are correct K-mers supporting its further extension). Stage 4, sequence refinement, evaluating whether chemical residue conversions (Gln to Glu or Asn to Asp) occur at given positions or whether mistakes were incorporated in the assembly process. All peptides are aligned onto the assembled sequence and per position evaluation is done. Rationale, remaining unconverted residues at specific positions reveal their identity prior to MAAH. Furthermore, peptide tags generated by enzymatic digestion can additionally be used by pTA for resolving chemical conversion ambiguities.