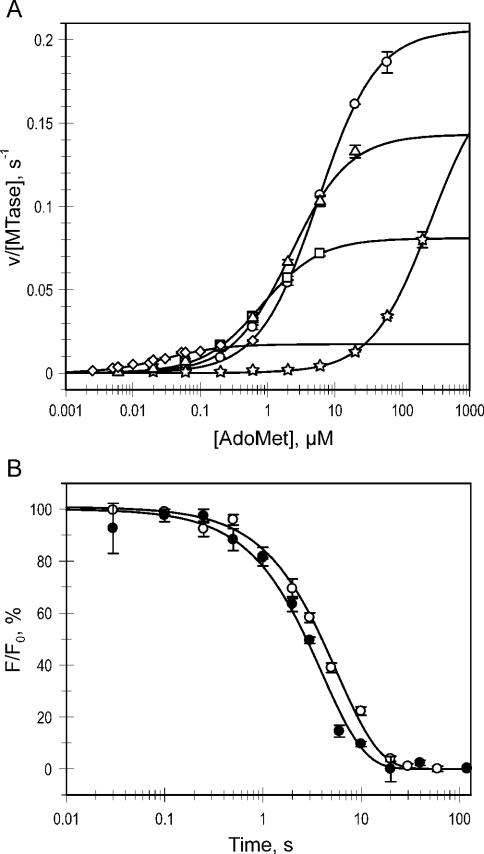
Figure 2.
Kinetic analysis of Trp41 mutants. (A) Steady-state velocities for the mutants (circles, W41G; triangles, W41A; squares, W41F; diamonds, WT; stars, W41P) were determined at constant saturating poly(dG–dC) and varied [methyl-3H]AdoMet concentrations and the data were fitted to a Michaelis–Menten equation to reveal and kcat (shown in Figure 3). (B) Single-turnover methylation reactions were performed by incubation of a biotin- and JOE-labeled 34mer DNA duplex with excess MTase and AdoMet in a rapid-quench-flow device. The DNA was immobilized and digested with R.Hin6I to release fluorescent fragments from unmethylated DNA. Fluorescence time courses (open circles, WT; filled circles, W41G) were fitted to a single exponential equation (solid lines) to reveal kchem.