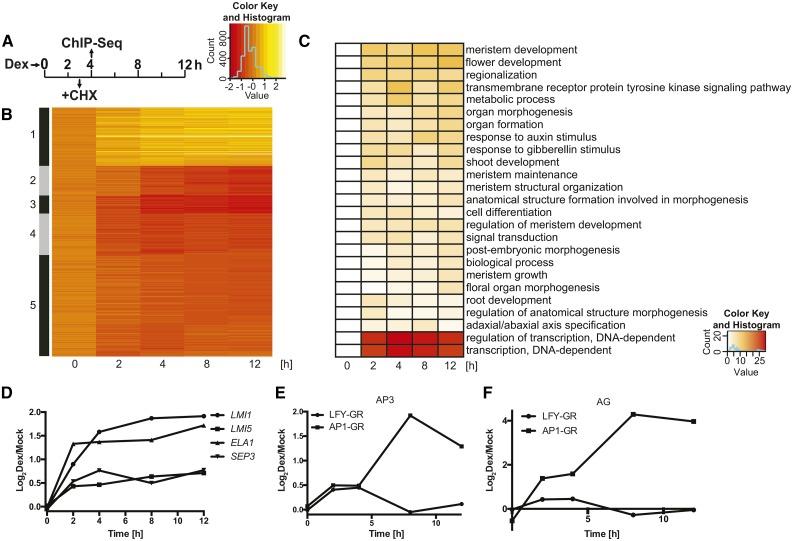
Figure 1.
Genes controlled by LFY in the absence of AP1/CAL function. A, Experimental set-up of the genome-wide analyses using inflorescence-like meristems from p35S:LFY-GR ap1 cal plants. CHX, cycloheximide; Dex, dexamethasone. B, K-means clustering (k = 5) of 669 DEGs identified in the p35S:LFY-GR ap1 cal time course experiment. Log2-transformed expression ratios (dexamethasone/mock) for the different time points (as indicated) were used for the analysis. C, Gene Ontology terms enriched among the DEGs at different time points (as indicated). Negative decadal logarithms of P values are shown. D, Response of selected LFY targets (as indicated) to an activation of LFY-GR in ap1 cal inflorescences. E and F, Response of AP3 (E) and AG (F) to an activation of LFY-GR and AP1-GR, respectively, in ap1 cal inflorescences. Data for AP1-GR were taken from Kaufmann et al. (2010). In D to F, log2-transformed fold-change values (dexamethasone/mock) from microarray experiments are shown.