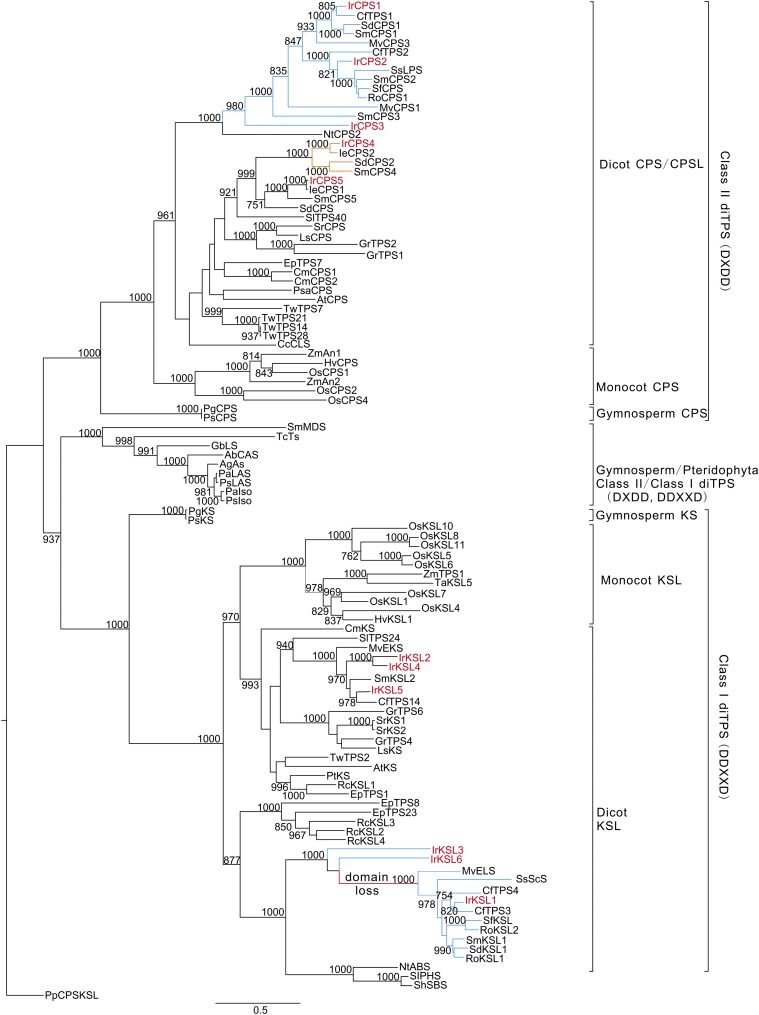
Figure 3.
Phylogeny of I. rubescens diterpene synthases. The maximum likelihood tree illustrates the phylogenetic relationship of I. rubescens diterpene synthases with 96 representative characterized diTPS (Supplemental Table S5). Numbers on branches indicate the bootstrap percentage values calculated from 1000 bootstrap replicates. Physcomitrella patens CPS/kaurene was used as an outgroup. Blue lines show diTPS genes from Lamiaceae involved in normal-CPP/LDPP-mediated diterpenoid metabolism and red lines show the loss of the N-terminal γ domain found in eudicot and monocot KSLs. Yellow lines show genes from Lamiaceae involved in the specialized ent-CPP/LDPP-related diterpenoid metabolism. Red-marked enzymes show diTPS from I. rubescens in this study.