GA regulates SL biosynthesis through the GA receptor GID1 and F-box protein GID2.
Abstract
Strigolactones (SLs) are a class of plant hormones that regulate diverse physiological processes, including shoot branching and root development. They also act as rhizosphere signaling molecules to stimulate the germination of root parasitic weeds and the branching of arbuscular mycorrhizal fungi. Although various types of cross talk between SLs and other hormones have been reported in physiological analyses, the cross talk between gibberellin (GA) and SLs is poorly understood. We screened for chemicals that regulate the level of SLs in rice (Oryza sativa) and identified GA as, to our knowledge, a novel SL-regulating molecule. The regulation of SL biosynthesis by GA is dependent on the GA receptor GID1 and F-box protein GID2. GA treatment also reduced the infection of rice plants by the parasitic plant witchers weed (Striga hermonthica). These data not only demonstrate, to our knowledge, the novel plant hormone cross talk between SL and GA, but also suggest that GA can be used to control parasitic weed infections.
Strigolactones (SLs) are a group of terpenoid lactones that regulate shoot branching outgrowth and root development in various plant species (Gomez-Roldan et al., 2008; Umehara et al., 2008; Koltai, 2011a; Ruyter-Spira et al., 2011; Seto et al., 2012). SLs are also exuded from roots into the rhizosphere as signaling molecules that stimulate the germination of root parasitic weeds and the branching of arbuscular mycorrhizal fungi (Cook et al., 1966; Akiyama et al., 2005). At present, two carotenoid cleavage dioxygenases (D10 and D17), a carotenoid isomerase (D27), Os01g0700900 (Os0900), and Os01g0701400 (Os1400) are known to be involved in the biosynthesis of SLs in rice (Oryza sativa; Lin et al., 2009; Cardoso et al., 2014; Zhang et al., 2014; Seto et al., 2014; Abe et al., 2014). The activities of SLs depend on D3 and D14: D3 encodes the F-box protein and D14 encodes SL receptor, which act in SL signaling. D53, which encodes a substrate of the SCFD3 complex, was recently reported to be a repressor of SL signaling in rice, and the degradation of D53 protein by SL, in cooperation with D14 and D3, is considered as a key event in SL signaling (Jiang et al., 2013; Zhou et al., 2013; Yao et al., 2016; de Saint Germain et al., 2016).
Gibberellin (GA) is a plant hormone that regulates many aspects of plant growth and development during the life cycle of plants (Yamaguchi, 2008). Plants defective in GA biosynthesis or signaling show characteristic phenotypes, including dwarfism, small dark green leaves, prolonged germination dormancy, root growth retardation, suppression of flowering, reduced seed production, and male sterility (Yamaguchi, 2008). Several factors involved in GA perception have been identified. In rice, GA binds to the GA receptor GID1, and GA binding promotes the interaction between GID1 and the transcriptional repressor DELLA protein, SLR1. GA-induced interaction of GID1 and SLR1 triggers the ubiquitination of SLR1 by the GID2 F-box protein, one of the components of an SCF-type E3 ubiquitin ligase that degrades SLR1 through the 26S proteasomal pathway (Ueguchi-Tanaka et al., 2005; Sasaki et al., 2003). GA-dependent loss of the repressor activity of DELLA is considered a key step in GA signaling (Ueguchi-Tanaka et al., 2008).
Root parasitic weeds, such as broomrape (Orobanche spp.) and witchers weed (Striga spp.), are harmful plants in sub-Saharan Africa, the Middle East, and Asia that maintain seed dormancy in the absence of host plant (Spallek et al., 2013). The germination mechanisms of Orobanche and Striga spp. are very similar. Both species require germination stimulants released from the host plant. SLs are the major group of germination stimulants effective for almost all Orobanche and Striga spp. It has been reported that approximately 300 million people are affected economically by Striga spp. in Africa, with estimated losses of $US 7 billion (Parker, 2009). Although many approaches to controlling these parasitic weeds have been explored, their success has been limited. One approach is the method called “suicidal germination”, which induces the germination of the parasites’ seeds in the absence of the host plants, because Striga and Orobanche spp. are obligate parasites. However, this approach is not cost effective for controlling these weeds expanded into a large area.
There is some cross talk between SLs and various hormones. For example, auxin, which regulates shoot branching and root morphology, acts both up- and downstream from SL signaling, in secondary growth and root hair elongation, respectively (Agusti et al., 2011; Koltai, 2011a). SL also inhibits auxin transport by reducing the accumulation of auxin efflux carrier component1 in xylem parenchyma cells (Crawford et al., 2010). Ethylene also affects root hair elongation by exerting epistatic effects on SLs (Kapulnik et al., 2011). Two reports have also directly demonstrated an interaction between SL and hormonal signaling. Wang et al. (2013) showed an interaction between the SL and brassinosteroid signaling pathways. BES1, an important transcriptional regulator in the brassinosteroid signaling pathway, interacts with MAX2, which is the ortholog of rice D3, and BES1 degradation is accelerated by SLs in Arabidopsis (Arabidopsis thaliana). Nakamura et al. (2013) reported an SL-dependent interaction between D14 and SLR1 in rice. This data suggests the existence of cross talk between the GA and SL signaling pathways. However, contradicting results have been reported in which SLs act independently of GAs in stimulating internode elongation in the pea (Pisum sativum; de Saint Germain et al., 2013). Therefore, there is as yet no physiological evidence of the existence of cross talk between the GA and SL signaling pathways, although Lo et al. (2008) reported that GA deficiency promotes tiller bud elongation in rice.
To identify, to our knowledge, a novel regulator of SL biosynthesis that controls the germination of root parasitic weeds in this study, we screened for chemicals that regulate SL biosynthesis. We found that GA is a regulator of SL biosynthesis, and that GA signaling controls the biosynthesis of SL by regulating the expression of SL biosynthesis genes. Moreover, GA-treated rice showed reduced Striga infection. Therefore, we demonstrate the cross talk between the GA and SL signaling pathways for the first time to our knowledge, and also present, to our knowledge, new insight into the management of root parasitic weeds.
RESULTS
GAs Regulate SL Biosynthesis
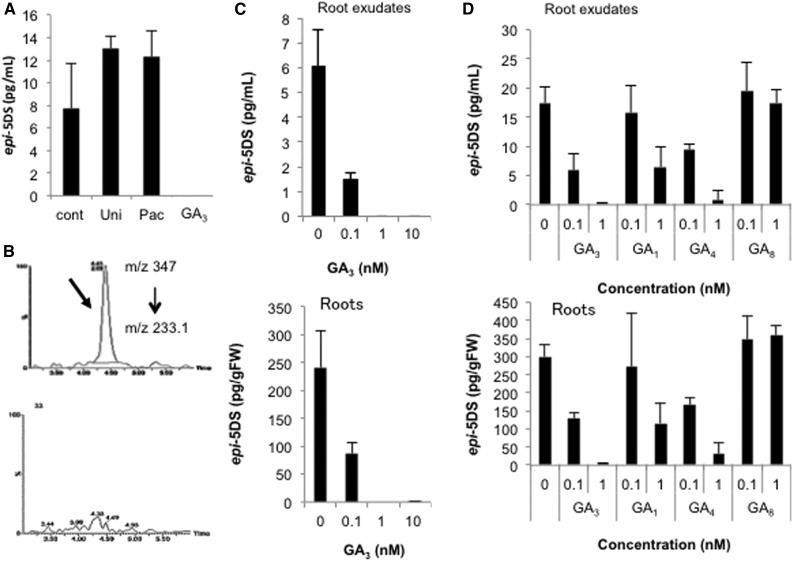
Because root parasitic weeds such as Striga and Orobanche germinate in response to SL in a concentration-dependent manner, low-SL-producing plants can be resistant to infection by these weeds (Jamil et al., 2012). In an attempt to find, to our knowledge, a novel SL biosynthesis regulator, we screened several chemicals, including plant hormones, plant growth regulators, and triazole derivatives previously constructed in our laboratory (Min et al., 1999; Sekimata et al., 2001, 2002). To evaluate their ability to regulate SL biosynthesis, we analyzed the levels of 2′-epi-5-deoxystriol (epi-5DS), an endogenous SL in rice, in root exudates, using liquid chromatography-tandem mass spectroscopy (LC-MS/MS). Because the SL levels in the root exudates of rice seedlings are elevated when inorganic P is depleted in the medium (Umehara et al., 2008), we examined the effects of these chemical treatments on SL levels under P-deficient conditions. With screening, we identified a plant hormone, GA3, that strongly reduced the levels of epi-5DS in the root exudates (Fig. 1A). Another SL (orobanchol) in the root exudates was nearly undetectable in the GA3-treated rice (Fig. 1B). The application of a GA biosynthesis inhibitor (1 μm uniconazole-P or 1 μm paclobutrazol) also slightly increased the levels of epi-5DS (Fig. 1A). To exclude the possibility that GA3 inhibits SL export and reduces the levels of epi-5DS in root exudates, we analyzed the endogenous epi-5DS levels in rice roots. Our data clearly indicate that GA3 strongly and dose-dependently reduces the levels of epi-5DS in both roots and root exudates in a concentration range of 0.1 to 10 nm (Fig. 1C). Moreover, the reduced levels of epi-5DS in a GA3-treated Lotus japonicus root cell culture suggested that GA3 also reduces the levels of SL in in vitro culture systems and other plant species (Supplemental Fig. S1).
Figure 1.
Effects of GA on SL levels in 2-week-old rice. A, Epi-5DS levels in root exudate of rice treated with GA3 and a GA biosynthesis inhibitor, detected with LC-MS/MS. B, Monitoring selected ions for orobanchol. Upper and lower panels show the chromatograms for the control and 10 nm GA3-treated root exudates, respectively. C, Epi-5DS levels in root exudates (upper) and roots (lower) of GA3-treated rice, determined with LC-MS/MS. D, Epi-5DS levels in root exudates (upper) and roots (lower) of rice treated with various GAs, determined with LC-MS/MS. Thirteen-d-old rice seedlings were treated for 1 d with chemicals. Data are means ± sd (n = 3). GA3, 10 nm GA3; Pac, 1 μm paclobutrazol; Uni, 1 μm uniconazole-P.
Many GAs are present in plants. In many cases, the intensity of GA responses is dependent on GA-GID1 binding affinity (Ueguchi-Tanaka et al., 2005). To examine the effects of various GAs on SL production, we analyzed the levels of epi-5DS in rice treated with active (GA1, GA3, and GA4) or inactive (GA8) GAs. The effects of GAs on SL production were generally consistent with the physiological activities of each GA (Fig. 1D; Nishijima et al., 1994). These results indicate that bioactive GAs reduce SL production.
Regulation of SL Biosynthesis via a GID-DELLA Signaling Pathway
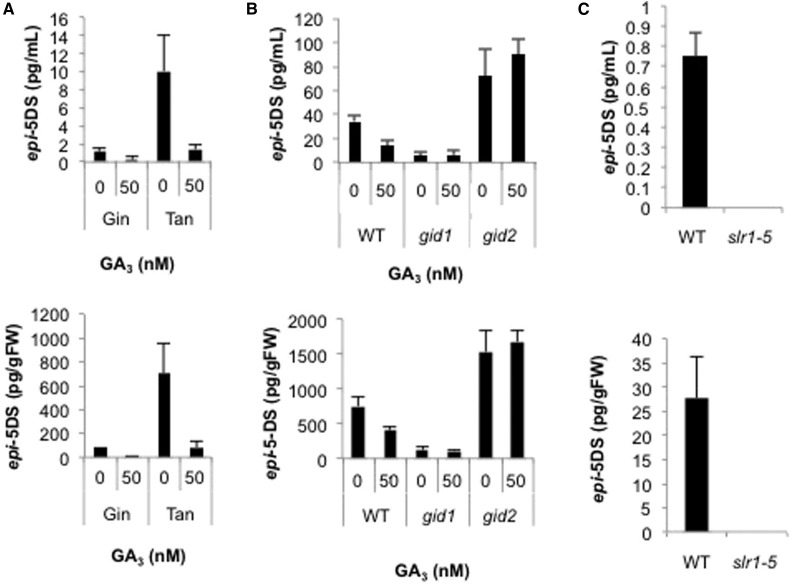
To investigate the mechanism underlying the regulation of SL biosynthesis by GA, the levels of epi-5DS were measured in the roots and root exudates of mutants defective in GA biosynthesis or signaling, with LC-MS/MS (Fig. 2). In the GA biosynthesis mutant, Tanginbozu, which shows a semidwarf phenotype, elevated levels of epi-5DS were detected in the roots and root exudates, and were suppressed after GA treatment (Fig. 2A). In GA-insensitive mutants gid1-3 and gid2-2, the level of epi-5DS did not change after GA treatment (Ueguchi-Tanaka et al., 2005; Sasaki et al., 2003). The level of epi-5DS was also higher in the gid2-2 mutant than in wild-type seedlings (Fig. 2B). SLR1 is a repressor of GA signaling and most GA-related responses are induced by the degradation of SLR1. One SLR1 mutant, slr1-5, is a constitutive GA response mutant. epi-5DS was undetectable in the roots and root exudates of the slr1-5 mutant (Fig. 2C). These results suggest that the regulation of SL production by GAs is caused by a GID-DELLA signaling pathway.
Figure 2.
Effects of GA on SL levels in GA biosynthesis and signaling mutants. Epi-5DS levels in root exudates (upper) and roots (lower) of GA3-treated rice, determined with LC-MS/MS. A, GA biosynthesis mutant (Tanginbozu). B, GA signaling mutants (gid1 and gid2). C, Constitutive GA signaling mutant (slr1-5). Thirteen-day-old rice seedlings were treated for 1 d with GA3. Data are means ± sd (n = 3). Gin, Ginbozu; Tan, Tanginbozu; WT, wild type.
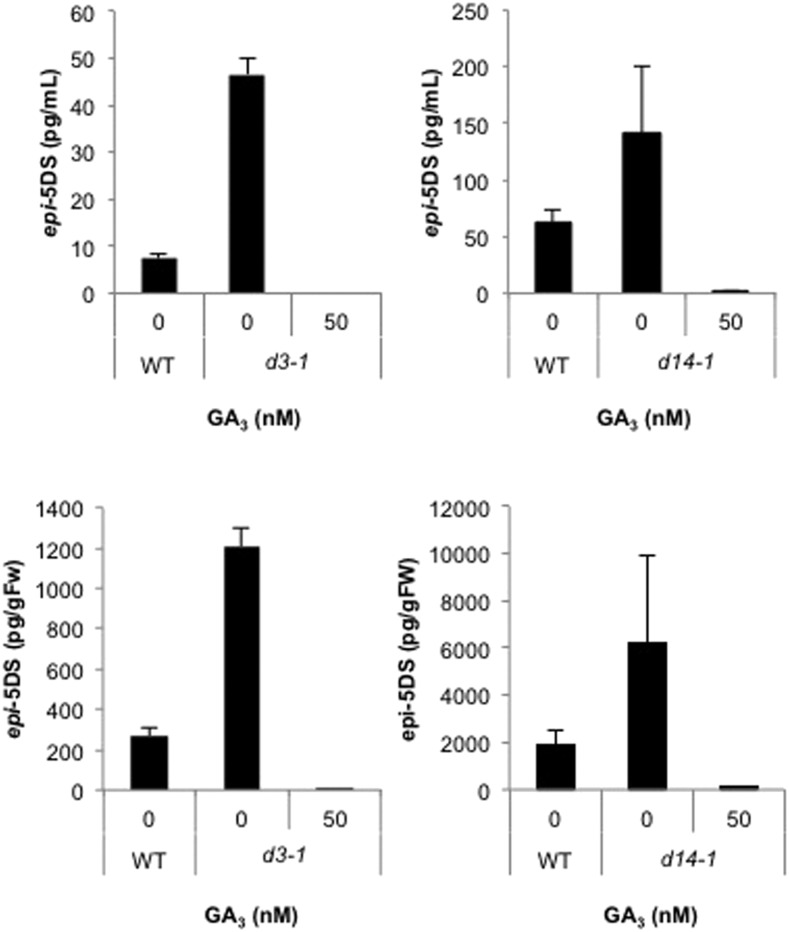
SL biosynthesis is tightly controlled by endogenous SL levels through the negative feedback regulation of D10 and D17 expression (Umehara et al., 2008). To determine whether the regulation of SL production by GAs depends on SL signaling, we tested the effects of GA on SL levels in an SL signaling mutant. Although the epi-5DS levels were higher in the loss of function mutants in SL signaling (d3-1 and d14-1) than in the wild type, presumably as a consequence of the feedback regulation of the SL pathway, GA3 reduced the levels of epi-5DS in both roots and root exudates (Fig. 3). These results indicate that the pathway through which GA3 regulates SL biosynthesis is independent of D3 and D14.
Figure 3.
Effects of GA on SL levels in SL signaling mutants. Epi-5DS levels in root exudates (upper) and roots (lower) of the d3-1 (left) and d14-1 (right) mutants, determined with LC-MS/MS. Thirteen-day-old rice seedlings were treated for 1 d with GA3. Data are means ± sd (n = 3). GA3, 10 nm GA3; WT, wild type.
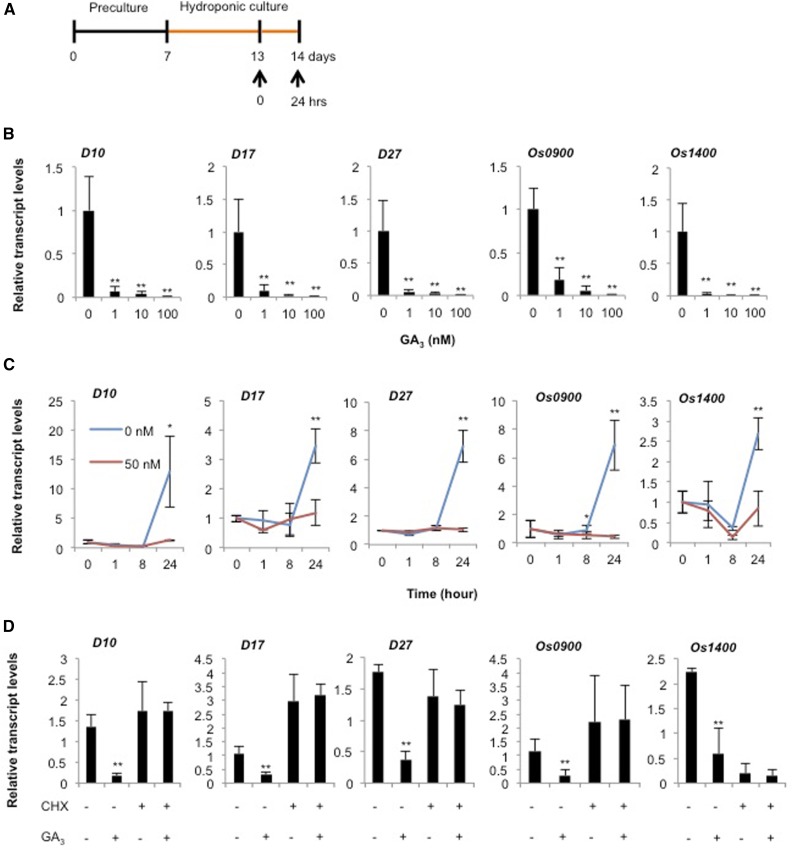
We used a quantitative reverse transcription PCR (qRT-PCR) analysis of the SL biosynthesis genes to clarify the mechanism underlying the regulation of SL biosynthesis by GA signaling (Fig. 4A). SLs are synthesized by OsD27, OsD10, OsD17, Os0900, and Os1400 in rice (Alder et al., 2012; Zhang et al., 2014; Supplemental Fig. S2). Our qRT-PCR analysis revealed that treatment with GA3 for 24 h reduced these transcript levels in the wild-type roots (Fig. 4B). A time-course analysis of the SL biosynthesis genes was performed in roots exposed to 50 nm GA3 for 0 to 24 h. All of the genes tested here showed similar expression patterns (Fig. 4C). In the control roots, the transcripts increased until 24 h, but remained low in the GA3-treated roots. This result suggests two possibilities: one is that GA treatment reduces transcript levels of SL biosynthesis genes, and the other is GA suppresses the up-regulation of SL biosynthesis genes and maintains the transcription at basal level. To clarify these hypotheses, we performed the expression analysis of SL biosynthesis genes in the time course shown in Supplemental Figure S3A. The transcript levels of SL biosynthesis genes were reduced by GA treatment (Supplemental Fig. S3B). This result indicates GA down-regulates SL production through mediating the expression levels of SL biosynthesis genes. Next, we estimated the effects of a protein synthesis inhibitor, cycloheximide (CHX), on the regulation of SL biosynthesis by GA signaling. As mentioned above, treatment with 50 nm GA3 reduced the transcript levels of the SL biosynthesis genes. On the other hand, CHX abolished the repression by GA on the transcript levels in comparison with the control (Fig. 4C). These results imply that the regulation of SL levels by GA signaling is attributable to the altered expression of the SL biosynthetic genes via de novo protein synthesis.
Figure 4.
Effects of GA on transcript levels of SL biosynthesis genes in roots. A, Schematic diagram showing the experimental conditions. Orange bar indicates a hydroponic culture without P. B, Transcript levels of SL biosynthesis genes in GA-treated rice roots. C, Time-course analysis of the expression of SL biosynthesis genes after treatment with 50 nm GA3. D, Effect of protein synthesis inhibitor (CHX) on the expression levels of SL biosynthesis genes after treatment with 50 nm GA3. Thirteen-d-old rice seedlings were transferred to new vials containing fresh media with or without chemicals for 1 d (B and D) or the indicated times (C). Data are means ± sd (n = 3). Asterisks (*) and (**) indicate signify differences from 0 nm GA3-treated plants (t test, P < 0.05 and P < 0.01, respectively). GA3, 10 nm GA3.
Effects of SL Treatment on GA-Deficient Mutant
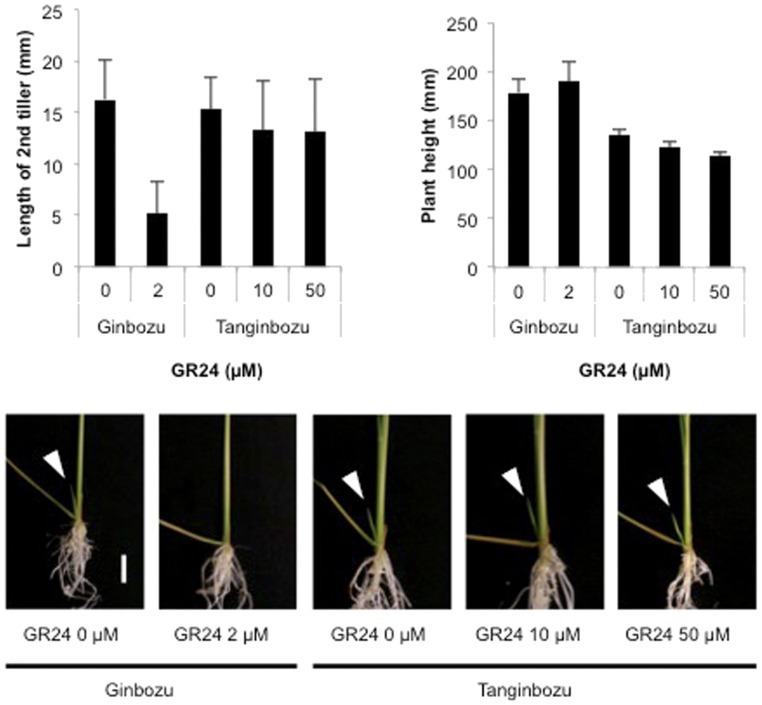
GA-deficient mutants show increased tiller bud outgrowth and dwarfism (Lo et al., 2008; Ito et al., 2010). To evaluate the effects of SL on GA-deficient rice, we used the GA-deficient mutant Tanginbozu. Tanginbozu displays dwarfism and greater tiller outgrowth in 5-week-old seedlings than the wild type (Ginbozu; Supplemental Fig. S4), although the length of the second tiller buds of Tanginbozu did not differ from that of Ginbozu in 2-week-old seedlings (Fig. 5). The length of the second tiller was suppressed in Ginbozu by the application of 2 μm GR24, a synthetic SL analog, whereas no response was observed in the length of the second tiller bud of Tanginbozu. Plant height did not differ between the control and the GR24-treated plants of either Ginbozu or Tanginbozu. This result suggests that GA is required for the regulation of tiller bud outgrowth by SL.
Figure 5.
Effects of GR24 on tiller bud growth in GA biosynthesis mutant. Second tiller length and plant height in 2-week-old rice. Data are means ± sd (n = 6). White arrowheads indicate second tillers. Scale bar: 1 cm. Gin, Ginbozu; Tan, Tanginbozu.
GA-Treated Rice Is Infected by Fewer Striga Plants
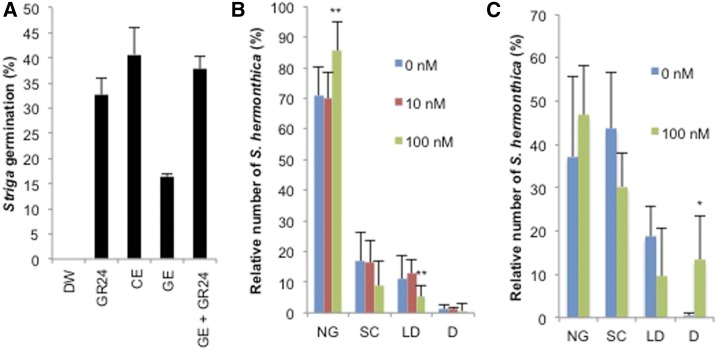
We have demonstrated that GA regulates SL biosynthesis. To explore the effects of GA treatment on the interaction of plants with root parasitic weeds, we performed Striga germination and infection assays. Consistent with the results of the epi-5DS level in GA-treated rice, the root exudates of rice seedlings treated with 50 nm GA contained less germination-stimulating activity than those of the control plants (Fig. 6A). Moreover, fewer seeds germinated in the vicinity of the roots of 100 nm GA-treated rice than in the vicinity of the control roots (Fig. 6B). As a result of the reduced germination frequency, statistically fewer Striga established parasitism on the 100 nm GA-treated rice. When germinated Striga seeds that had been stimulated with strigol were incubated with 100 nm GA-treated rice, there was no significant difference in the frequency of successful parasitism between the control and 100 nm GA-treated rice plants (Fig. 6C). This result indicates that GA treatment does not affect the infectious processes except for germination process.
Figure 6.
Effects of GA on Striga germination and infection. A, Striga germination rate after treatment with culture medium. Data are means ± sd (n = 3). DW, distilled water; GR24, 1 μm GR24; CE, culture medium for 0 nm GA3-treated rice; GE, culture medium for 100 nm GA3-treated rice. B and C, Ratio of Striga plants at each developmental stage 3 weeks after the inoculation of SL-treated (C) or untreated seeds (B). Concentrations in (B) and (C) indicate the GA concentrations of GA-treated rice. Data are means ± sd; B, n = 8 to 10; C, n = 2 to 3. Asterisks (* and **) indicate significantly different from untreated rice (t test, P < 0.05 and P < 0.01, respectively). NG, no germination; SC, penetration successful and seed coat remained attached; LD, leaf developed after the establishment of parasitism; D, died after penetration.
DISCUSSION
SLs are important phytohormones required for plant growth and development. Because SLs are known to be involved in the regulation of diverse physiological phenomena, including shoot branching, root development, and leaf senescence, they are thought to be involved in physiological interactions with various hormones and environmental cues (Crawford et al., 2010; Dun et al., 2012; Kapulnik et al., 2011; Ha et al., 2014; de Jong et al., 2014; Mayzlish-Gati et al., 2012; Tsuchiya et al., 2010). However, the physiological cross talk between SL and GA has not been determined, despite their molecular interaction, in which the putative SL receptor, D14, interacts with the GA signaling repressor, SLR1 (Nakamura et al., 2013). Here, we provide evidence of the interaction between these two hormones, by showing that GA signaling regulates SL biosynthesis.
GA signaling negatively regulates the endogenous levels of SLs. Importantly, the application of an inactive GA metabolite (GA8) to wild-type plants or active GA to GA signaling mutants (gid1-3 and gid2-2) did not induce this regulation, whereas the application of active GA to the wild type and a GA biosynthesis mutant (Tanginbozu) reduced their levels of SLs (Figs. 1 and 2). We also detected no SL in a constitutive GA response mutant (slr1-5). These results indicate that the regulation of endogenous SL level by GA signaling depends on the activity of the DELLA protein. The repressive activity of GA on SL biosynthesis did not correlate with the binding activity of GA to GID1 (Fig. 1D; Ueguchi-Tanaka et al., 2005). GA4 shows the highest affinity for GID1. However, whereas GA1 and GA4 are inactivated by GA 2-oxidase in plants, GA3 is not oxidized by GA 2-oxidase (Nakayama et al., 1990). These facts explain the inconsistency between the repressive activity on SL biosynthesis and the binding activity on GID1. The increased levels of SL in gid2-2 are similar to those in Tanginbozu, suggesting that a reduction in GA signaling induces an increase in the level of SLs. Interestingly, however, the level of SL was lower in the gid1-3 mutant than in the wild type (Fig. 2, A and B), whereas both the gid1-3 and gid2-2 mutants were insensitive to GA and had similar phenotypes. These data suggest that a GID1-independent GA signaling pathway could be present, similar to the GID2-independent GA signaling pathway reported by Ueguchi-Tanaka et al. (2008), although this regulatory mechanism is still unclear. Alternatively, as GID1 and D14 competitively bind to SLR1, by the loss of function of GID1 binding to GA, D14 can dominantly bind to SLR1 and suppress SLR1 function. As a result, in the gid1 mutant 5DS, the level decrease could be due to feedback regulation similar to in that in the slr1 mutant.
SL biosynthesis is positively and negatively regulated by various hormones and environmental cues (Yoneyama et al., 2007a, 2007b; Hayward et al., 2009; Koltai et al., 2011b; López-Ráez et al., 2010). Phosphate is a negative regulator of SL biosynthesis and its regulatory activity alters the expression of the SL biosynthetic genes (Umehara et al., 2010). A previous gene expression analysis indicated that P deficiency increased the transcription levels of D10, D17, D27, Os0900, Os01g0701500, and Os02g0221900 (Umehara et al., 2010), whereas GA signaling regulated the expression of the Os1400 gene, as well as D10, D17, D27, and Os0900 (Fig. 2). These results suggest that GA signaling regulates the expression of the SL biosynthesis genes through a different pathway from P signaling. However, CHX treatment reduced the transcription level of Os1400, whereas the other SL biosynthesis genes (D10, D17, D27, and Os0900) were hardly affected by CHX treatment, suggesting that the regulatory mechanisms of Os1400 expression by GA signaling were different from those of the other SL biosynthesis genes.
In the time course experiment, the expression levels of SL biosynthesis genes were increased at 24 h (Fig. 4C). We did not exchange hydroponic culture media from d 7 to d 13 and transferred rice seedlings into fresh media at d 13 (Fig. 4A). Removal of the accumulated SLs from media might lead the up-regulation of SL biosynthesis genes.
Tanginbozu showed the GR24-insensitive phenotype in tiller bud length (Fig. 5). In Tanginbozu, the endogenous level of epi-5DS was higher than that in Ginbozu (Fig. 2A). In addition, d10-1 mutant is more sensitive to GR24 than the wild type (Umehara et al., 2008). Therefore, the result that Tanginbozu displayed a GR24-insensitive phenotype suggests the possibility that the SL signal is saturated with the accumulated SL in Tanginbozu. However, the overexpression of GA 2-oxidase also positively regulated the number of tillers, indicating the importance of GA signaling in shoot outgrowth in rice (Lo et al., 2008). Further physiological studies may contribute to the elucidation of the mechanism of shoot outgrowth.
In many parts of the world, Striga and Orobanche are serious agricultural pests. Reducing SL biosynthesis is one way to control these pests and low-SL-producing plants are actually resistant to root parasitic weed infections (Jamil et al., 2012; Umehara et al., 2008). In this study, we have shown that GA-treated rice was infected by fewer Striga plants than the control rice, at least under experimental conditions. Therefore, tissue-specific modification of GA signaling can be used to control root parasitic weed infection.
MATERIALS AND METHODS
Plant Material
The wild-type rice varieties (Oryza sativa) used in this study were Shiokari (d3-1 and d10-1; Ishikawa et al., 2005), Nipponbare (d10-2; Arite et al., 2007), Taichung 65 (slr1-5, gid1-3, and gid2-2; Ueguchi-Tanaka et al., 2008), and Ginbozu (Tanginbozu; Itoh et al., 2004).
Growth Conditions
Rice seedlings were grown hydroponically as described in Umehara et al. (2008). Surface-sterilized rice seeds were incubated in sterile water at 25°C for 2 d in the dark. For the analysis of SL, germinated seeds were transferred into hydroponic culture medium (Kamachi et al., 1991) solidified with 0.7% agar and cultured at 25°C for 6 d under fluorescent white light with a photoperiod of 14 h light/10 h dark. Each seedling was transferred to a glass vial containing 12 mL of sterilized hydroponic culture solution and grown under the same conditions for 6 d. The seedlings were then transferred to a new glass vial containing the culture solution, with or without chemicals, for 1 d. For the tillering experiment, the germinated seeds were transferred into hydroponic culture medium with 0.7% agar, with or without chemicals, under the same conditions. Each seedling was transferred to a new identical vial with or without chemicals. The hydroponic solution was refreshed every 3 d.
LC-MS/MS Analysis
The SL analysis was performed according to a previously described method (Umehara et al., 2008). Briefly, the hydroponic culture medium was extracted twice with ethyl acetate after the addition of d6-epi-5DS (200 pg) as the internal standard. The organic layer was dried and dissolved in 1 mL of ethyl acetate:n-hexane (15:85). The solutions were loaded onto a Sep-Pak silica 1 mL cartridge (Waters), washed twice with the same solution, eluted three times with ethyl acetate:n-hexane (35:65), and concentrated in vacuo. The roots were homogenized in acetone containing d6-epi-5DS. The filtrates were dried and dissolved in water. The solutions were extracted twice with ethyl acetate, dried, and dissolved in 10% acetone. The extracts were loaded onto Oasis HLB 3 mL cartridges (Waters), washed twice with water, eluted twice with acetone, and dried in vacuo. The concentrates were dissolved in 1 mL of ethyl acetate:n-hexane (15:85) and loaded onto Sep-Pak silica 1 mL cartridges, washed, eluted, and concentrated in the same way.
The SL-containing fractions were dissolved in 50% acetonitrile and subjected to an LC-MS/MS analysis in a system consisting of a quadruple/time-of-flight tandem mass spectrometer (Triple TOF 5600 system; AB Sciex) and an ultra-high-performance liquid chromatograph (Nexera; Shimadzu) equipped with a reversed-phase column (Acquity UPLC BEH-C18, 2.1 × 50 mm, 1.7 μm; Waters). The mobile phase was changed from 30% acetonitrile containing 0.05% acetic acid, to 40% acetonitrile at 5 min and to 70% acetonitrile at 10 min after injection, at a flow rate of 0.2 mL/min. The parent ions (m/z) were 331.2 for unlabeled epi-5DS, 337.2 for labeled epi-5DS, and 347.2 for orobanchol. The samples were quantified using fragment ions 234.13 for epi-5DS and 240.13 for d6-epi-5DS.
Striga Germination and Infection Assays
The Striga germination and infection assays were performed as described by Sugimoto and Ueyama (2008) and Umehara et al. (2008), respectively. For the infection assay, 1-week-old rice seedlings were transferred to root-observing rhizotron chambers (140 mm × 100 mm square petri dishes filled with rockwool and nylon mesh) supplied with 50 mL of nutrient solution with low P (1 mm NH4NO3, 0.06 mm NaH2PO4, 0.3 mm K2SO4, 0.3 mm CaCl2, 0.4 mm MgCl2, 45 μm Fe-EDTA with micronutrients; Makino et al., 1983) with or without GA3, and grown for 2 weeks in a greenhouse with a 12:12 h photoperiod (170 to 450 μmol/m2/s) with a day/night temperature cycle of 28°C/20°C. The Striga seeds were preconditioned on moist glass-fiber filter papers (GF/A; Whatman) at 26°C in the dark for 2 weeks, and treated with or without 10 nm strigol for 5 h in the dark. After they were rinsed with excess water, approximately 50 parasite seeds were carefully placed along the rice roots and the rhizotrons were incubated under the growth conditions described above. The germination, infection, and developmental status of the Striga plants were evaluated after cocultivation for 4 weeks (Yoshida and Shirasu, 2009).
Gene Expression Analysis
Total RNA was extracted from the roots using Plant RNA Purification Reagents (Invitrogen). cDNA was synthesized with the PrimeScript RT Reagent Kit with gDNA eraser (Takara Bio). qRT-PCR was performed on a Thermal Cycler Dice Real Time System using SYBR Premix Ex Taq (Takara Bio). The specific primers used for qRT-PCR are listed in the Supplemental Table.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers D10 (Os01g0746400), D17 (Os04g0550600), D27 (Os11g0587000), Os0900 (Os01g0700900) and Os1400 (Os01g0701400).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of GA3 on SL levels of L. japonicus root culture.
Supplemental Figure S2. SL biosynthesis pathway in rice.
Supplemental Figure S3. Effects of GA on transcript levels of SL biosynthesis genes in roots.
Supplemental Figure S4. Tiller number and plant height of GA biosynthesis mutant (Tanginbozu).
Supplemental Table S1. List of primers.
Supplementary Material
Footnotes
Articles can be viewed without a subscription.
This work was supported in part by grants from Core Research for Evolutional Science and Technology (to T.A.) and The Science and Technology Research Promotion Program for Agriculture, Fisheries and Food Industry (to T.A.).
References
- Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Kim HI, Yoneyama K, Xie X, Ohnishi T, et al. (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci USA 111: 18084–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, Greb T (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA 108: 20242–20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Cardoso C, Zhang Y, Jamil M, Hepworth J, Charnikhova T, Dimkpa SO, Meharg C, Wright MH, Liu J, Meng X, et al. (2014) Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proc Natl Acad Sci USA 111: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190 [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913 [DOI] [PubMed] [Google Scholar]
- de Jong M, George G, Ongaro V, Williamson L, Willetts B, Ljung K, McCulloch H, Leyser O (2014) Auxin and strigolactone signaling are required for modulation of Arabidopsis shoot branching by nitrogen supply. Plant Physiol 166: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C, et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Ligerot Y, Dun EA, Pillot JP, Ross JJ, Beveridge CA, Rameau C (2013) Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol 163: 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Ha CV, Leyva-González MA, Osakabe Y, Tran UT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Yamaguchi S, Dong NV, et al. (2014) Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci USA 111: 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Ito S, Kitahata N, Umehara M, Hanada A, Kato A, Ueno K, Mashiguchi K, Kyozuka J, Yoneyama K, Yamaguchi S, Asami T (2010) A new lead chemical for strigolactone biosynthesis inhibitors. Plant Cell Physiol 51: 1143–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Jamil M, Charnikhova T, Houshyani B, van Ast A, Bouwmeester HJ (2012) Genetic variation in strigolactone production and tillering in rice and its effect on Striga hermonthica infection. Planta 235: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Mae T, Ojima K (1991) A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiol 96: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H (2011) Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J Exp Bot 62: 2915–2924 [DOI] [PubMed] [Google Scholar]
- Koltai H. (2011a) Strigolactones are regulators of root development. New Phytol 190: 545–549 [DOI] [PubMed] [Google Scholar]
- Koltai H, Cohen M, Chesin O, Mayzlish-Gati E, Bécard G, Puech V, Ben Dor B, Resnick N, Wininger S, Kapulnik Y (2011b) Light is a positive regulator of strigolactone levels in tomato roots. J Plant Physiol 168: 1993–1996 [DOI] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, Wang Y (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Kohlen W, Charnikhova T, Mulder P, Undas AK, Sergeant MJ, Verstappen F, Bugg TD, Thompson AJ, Ruyter-Spira C, Bouwmeester H (2010) Does abscisic acid affect strigolactone biosynthesis? New Phytol 187: 343–354 [DOI] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K (1983) Photosynthesis and ribulose 1,5-bisphosphate carboxylase in rice leaves: changes in photosynthesis and enzymes involved in carbon assimilation from leaf development through senescence. Plant Physiol 73: 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayzlish-Gati E, De-Cuyper C, Goormachtig S, Beeckman T, Vuylsteke M, Brewer PB, Beveridge CA, Yermiyahu U, Kaplan Y, Enzer Y, et al. (2012) Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol 160: 1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min YK, Asami T, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (1999) New lead compounds for brassinosteroid biosynthesis inhibitors. Bioorg Med Chem Lett 9: 425–430 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K, Shi X, Ito E, Ito S, Park SH, et al. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 4: 2613. [DOI] [PubMed] [Google Scholar]
- Nakayama I, Miyazawa T, Kobayashi M, Kamiya Y, Abe H, Sakurai A (1990) Effects of a new plant growth regulator prohexadione calcium (BX-112) on shoot elongation caused by exogenously applied gibberellins in rice (Oryza sativa L.) seedlings. Plant Cell Physiol 31: 195–200 [Google Scholar]
- Nishijima T, Koshioka M, Yamazaki H (1994) Use of several gibberellin biosynthesis inhibitors in sensitized rice seedling bioassays. Biosci Biotechnol Biochem 58: 572–573 [Google Scholar]
- Parker C. (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci 65: 453–459 [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, Verstappen F, Bouwmeester H (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, Matsuoka M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Sekimata K, Han SY, Yoneyama K, Takeuchi Y, Yoshida S, Asami T (2002) A specific and potent inhibitor of brassinosteroid biosynthesis possessing a dioxolane ring. J Agric Food Chem 50: 3486–3490 [DOI] [PubMed] [Google Scholar]
- Sekimata K, Kimura T, Kaneko I, Nakano T, Yoneyama K, Takeuchi Y, Yoshida S, Asami T (2001) A specific brassinosteroid biosynthesis inhibitor, Brz2001: evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta 213: 716–721 [DOI] [PubMed] [Google Scholar]
- Seto Y, Kameoka H, Yamaguchi S, Kyozuka J (2012) Recent advances in strigolactone research: chemical and biological aspects. Plant Cell Physiol 53: 1843–1853 [DOI] [PubMed] [Google Scholar]
- Seto Y, Sado A, Asami K, Hanada A, Umehara M, Akiyama K, Yamaguchi S (2014) Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc Natl Acad Sci USA 111: 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T, Mutuku M, Shirasu K (2013) The genus Striga: a witch profile. Mol Plant Pathol 14: 861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Ueyama T (2008) Production of (+)-5-deoxystrigol by Lotus japonicus root culture. Phytochemistry 69: 212–217 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P (2010) A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol 6: 741–749 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M (2008) Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X (2013) Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 27: 681–688 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Yoneyama K, Takeuchi Y, Sekimoto H (2007a) Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K (2007b) Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227: 125–132 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shirasu K (2009) Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol 183: 180–189 [DOI] [PubMed] [Google Scholar]
- Zhang Y, van Dijk AD, Scaffidi A, Flematti GR, Hofmann M, Charnikhova T, Verstappen F, Hepworth J, van der Krol S, Leyser O, et al. (2014) Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat Chem Biol 10: 1028–1033 [DOI] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.