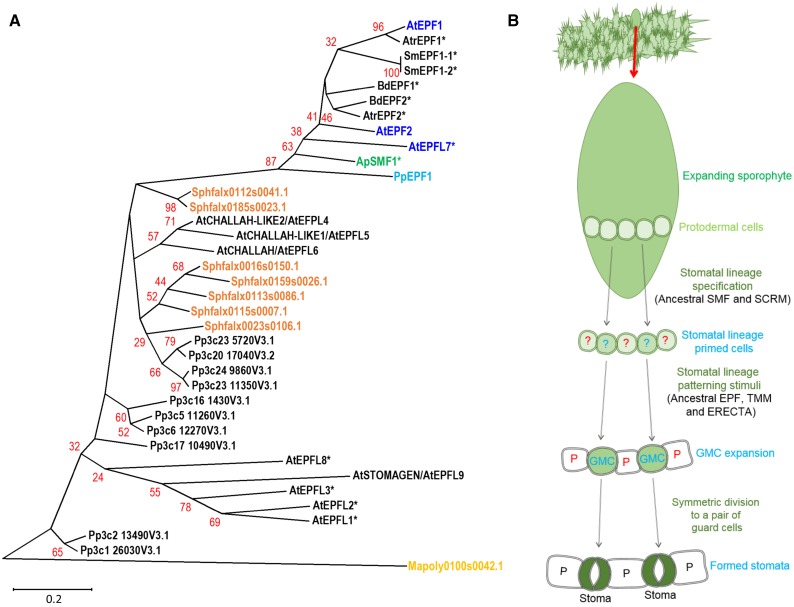
Figure 4.
EPF phylogeny of stomata- and non-stomata-bearing land plants and a model for stomatal development in the ancestor of stomata-bearing land plants. A, Phylogeny of EPF peptide sequences across the land plant kingdom. Stomatal and putative stomatal genes or closest equivalents in the liverwort M. polymorpha (yellow), the mosses S. fallax (orange) and P. patens (light blue), the hornwort A. punctatus (green), and the angiosperm Arabidopsis (dark blue) are marked to illustrate in which of these taxa the EPF stomatal patterning gene may be present. Starred genes (*) have not had their functions experimentally determined. The majority of sequences used to construct the phylogeny were obtained by BLAST comparison of the PpEPF1 amino acid sequence against the genomes of Mapoly, M. polymorpha; Sphfalx, S. fallax; Pp, P. patens; Sm, S. moellendorffii; Atr, Amborella trichopoda; Bradi, Brachypodium distachyon; and AT, Arabidopsis. Additional P. patens genes with more limited homology were added based on Caine et al. (2016). For simplicity, non-stomata-associated EPFs from vascular land plants have been omitted from the tree. The A. punctatus ApEPF1 partial sequence is based on Szövényi et al. (2015). Alignment and phylogenetic trees were prepared as described for Figure 2. Arabidopsis EPFL1-6 and 8-9 are included to highlight sequence relationships identified in (Caine et al., 2016). B, A model for stomatal development on the sporophyte of early evolving plants. Stomata precursors may have been specified via activity of an ancestral SMF-SCRM heterodimer. To ensure appropriate spacing of GMCs, cell signaling occurred via an ancestral patterning module (EPF, TMM, and ERECTA), followed by a final symmetric division leading to the formation of mature stomata.