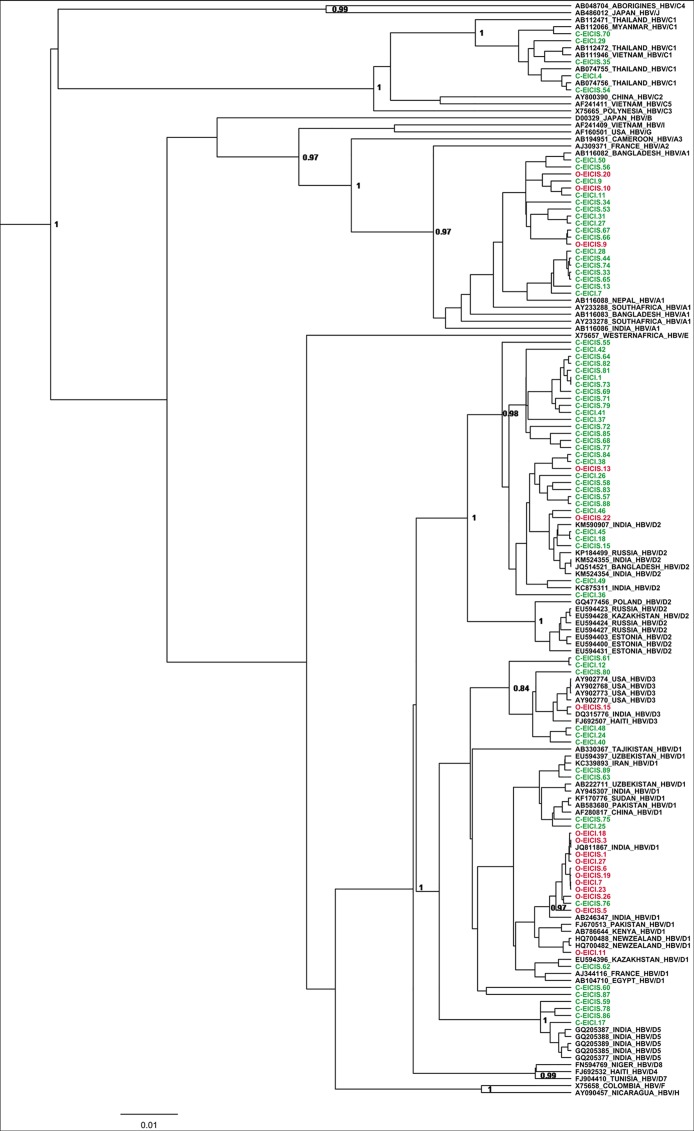
Fig 3. Phylogenetic analysis of occult and chronic HBV strains circulating among HIV-infected individuals in eastern India.
The phylogenetic relatedness was analyzed by means of Bayesian Markovchain Monte Carlo (MCMC) approach using a chain length of 50,000,000 and sampling in every 5000th generation and was based on the small surface (S) gene (nt 155–835 from EcoRI site). The occult HBV isolates (represented in red and labelled with O-) and the chronic HBV isolates (represented in green and labelled with C-) were analyzed with respect to GenBank reference sequences (represented in black) which are designated by their respective accession numbers along with their HBV genotypes/subgenotypes and country of origin. Posterior probabilities ≥0.9 are shown. HBV/D was the predominant genotype both among chronic (68.1%) and occult (82.4%) subjects, with HBV subgenotype D1 being widespread in OBI (64.7%) and D2 in chronic infection (42%).