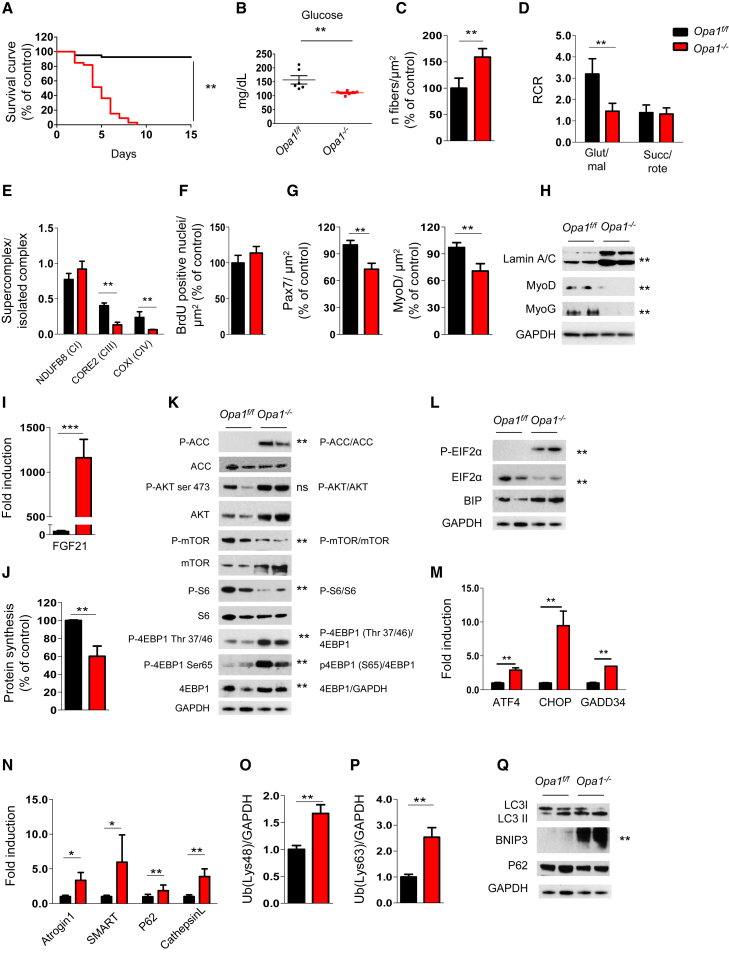
Figure 2.
OPA1 Deletion Induces a Lethal Phenotype Characterized by Mitochondrial Dysfunction, Reduction of Myogenic Stem Cells, Decreased Protein Synthesis, and Activation of Protein Breakdown
(A) Kaplan-Meier survival curve of Opa1f/f and Opa1−/− littermates (n = 37 for each group) indicates that muscle-specific OPA1 deletion results in lethality by post-natal day 9. ∗∗p < 0.01.
(B) Blood glucose levels in control and knockout mice (Opa1f/f n = 6; Opa1−/− n = 7). Data represent mean ± SEM, ∗∗p < 0.01.
(C) Analysis of the number of fibers normalized per muscle area (μm2). Data represent mean ± SEM for n = 6, ∗∗p < 0.01.
(D) Respiratory control ratio (RCR) of mitochondria energized with 5 mM/2.5 mM glutamate/malate (GLU/MAL) or 10 mM succinate (SUCC/ROTE). Data represent average ± SEM of three independent experiments (n = 9 per each condition), ∗∗p < 0.01.
(E) Densitometric quantification of Blue Native-PAGE of respiratory chain supercomplexes (RCS) that were normalized for individual respiratory chain complexes. Data represent mean ± SEM, n = 3, ∗∗p < 0.01.
(F) BrdU incorporation in Opa1f/f and Opa1−/− muscles. Data represent mean ± SEM, n = 4.
(G) Pax7-positive muscle stem cells and MyoD-positive myoblast were revealed by immunohistochemistry and normalized for muscle area. Data are mean ± SEM for n = 4, ∗∗p < 0.01.
(H) Representative immunoblot of muscle homogenates of three independent experiments. Lamin A/C, MyoD, and myogenin (MyoG) were normalized for GAPDH expression. Statistical significance after densitometric analysis is indicated on the right, ∗∗p < 0.01.
(I) Quantitative RT-PCR of FGF21 expression in Opa1−/− muscles. Data represent mean ± SEM, ∗∗∗p < 0.001, n = 4.
(J) Quantification of puromycin incorporation in skeletal muscles. In vivo SUnSET technique demonstrates a significant reduction of protein synthesis in Opa1−/− muscles. Data are mean ± SEM of three independent experiments (Opa1f/f n = 5; Opa−/− n = 7), ∗∗p < 0.01.
(K) Immunoblots of protein extracts from newborn muscles for the indicated antibodies. Representative immunoblots of three independent experiments are shown. Statistical analysis of the densitometric ratio is indicated on the right, n = 7 per each, ns: not significant, ∗∗p < 0.01.
(L and M) Representative immunoblots (L) and quantitative RT-PCR (M) showing activation of the UPR pathway in Opa1−/−. Data are mean ± SEM of three independent experiments, ∗∗p < 0.01.
(N) Quantitative RT-PCR analysis of FoxO-target genes. Data are mean ± SEM for n = 6, ∗∗p < 0.01.
(O and P) Densitometric quantification of lysine 48 (Lys48; O) and lysine 63 (Lys63; P) poly-ubiquitinated proteins from controls and Opa1−/− muscles. Data are mean ± SEM, n = 4, ∗∗p < 0.01.
(Q) Representative immunoblot of three independent experiments of autophagy-related proteins. Data are mean ± SEM, ∗∗p < 0.01.