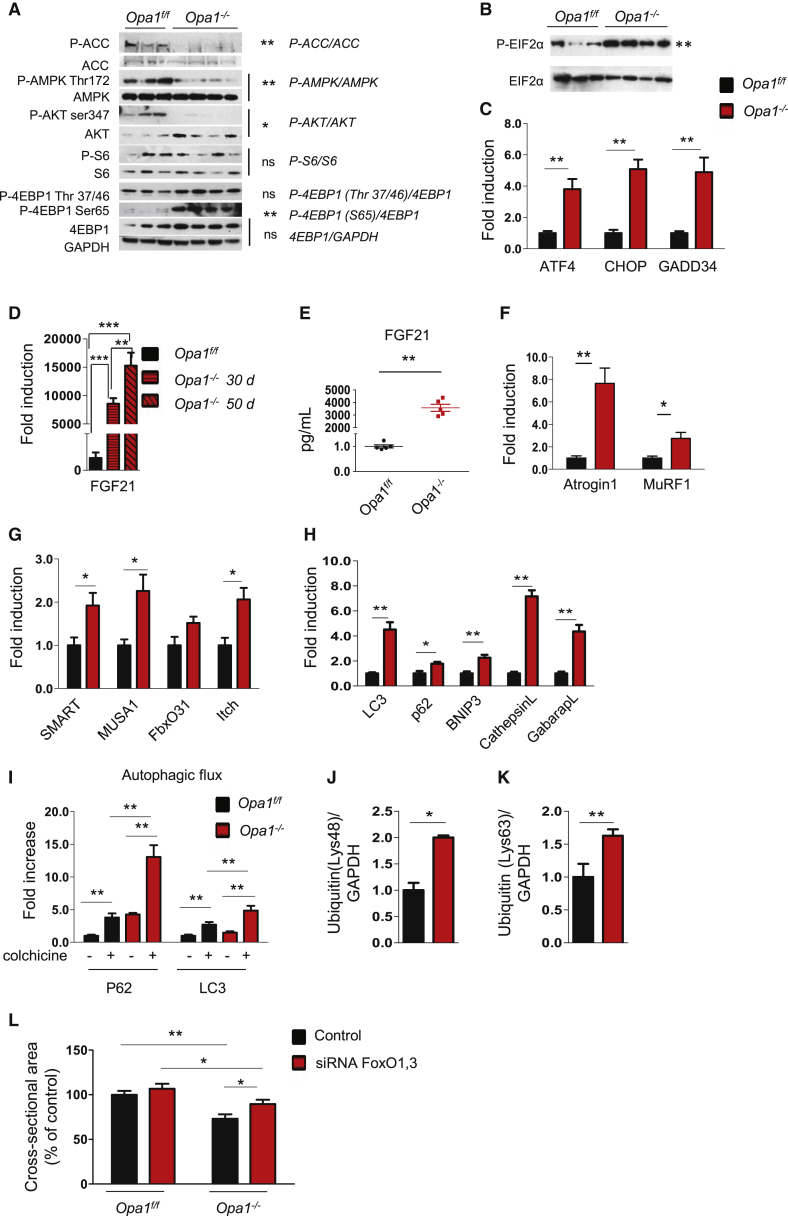
Figure 5.
Acute OPA1 Inhibition Does Not Affect Protein Synthesis but Triggers an Atrophy Program
(A) Total protein extracts from adult muscles were immunoblotted with the indicated antibodies. Representative immunoblots of two independent experiments are shown. Statistical significance after densitometry is indicated on the right. Data are mean ± SEM, ∗p < 0.05, ∗∗p < 0.01.
(B) p-eIF2α and total eIF2α immunoblots of muscle homogenates of three independent experiments. Statistical significance after densitometry is indicated on the right. Data are mean ± SEM, ∗∗p ≤ 0.01.
(C) Quantitative RT-PCR of genes downstream of UPR. Data are mean ± SEM, n = 5, ∗∗p < 0.01.
(D) Quantitative RT-PCR of FGF21 in OPA1-deficient muscles. Data are mean ± SEM, n = 4, ∗∗p < 0.01, ∗∗∗p < 0.001.
(E) FGF21 plasma levels in Opa1−/− mice were determined by ELISA (n = 6). ∗∗p < 0.01.
(F–H) Quantitative RT-PCR of several atrophy-related genes belonging to the ubiquitin proteasome (F and G) and autophagy-lysosome systems (H). Data are mean ± SEM, n = 5, ∗p < 0.05, ∗∗p < 0.01.
(I) Autophagy flux is increased in basal condition in OPA1-deficient muscles. Inhibition of autophagosome-lysosome fusion by colchicine treatment induces higher increase of p62 and LC3II band in OPA1 null than control muscles. Data are mean ± SEM, n = 5, ∗∗p ≤ 0.01.
(J and K) Densitometric quantification of lysine 48 (Lys48) and of lysine 63 (Lys63) poly-ubiquitinated proteins of muscle extracts from Opa1f/f and Opa1−/−. Data are mean ± SEM, n = 8, ∗p < 0.05, ∗∗p ≤ 0.01.
(L) Opa1−/− mice and Opa1f/f were transfected in vivo with scramble or siRNA against FoxO1/3. Inhibition of FoxOs by RNAi prevented muscle atrophy in Opa1−/− muscles. n = 6 per each group. Data are mean ± SEM, ∗p < 0.05, ∗∗p < 0.01.