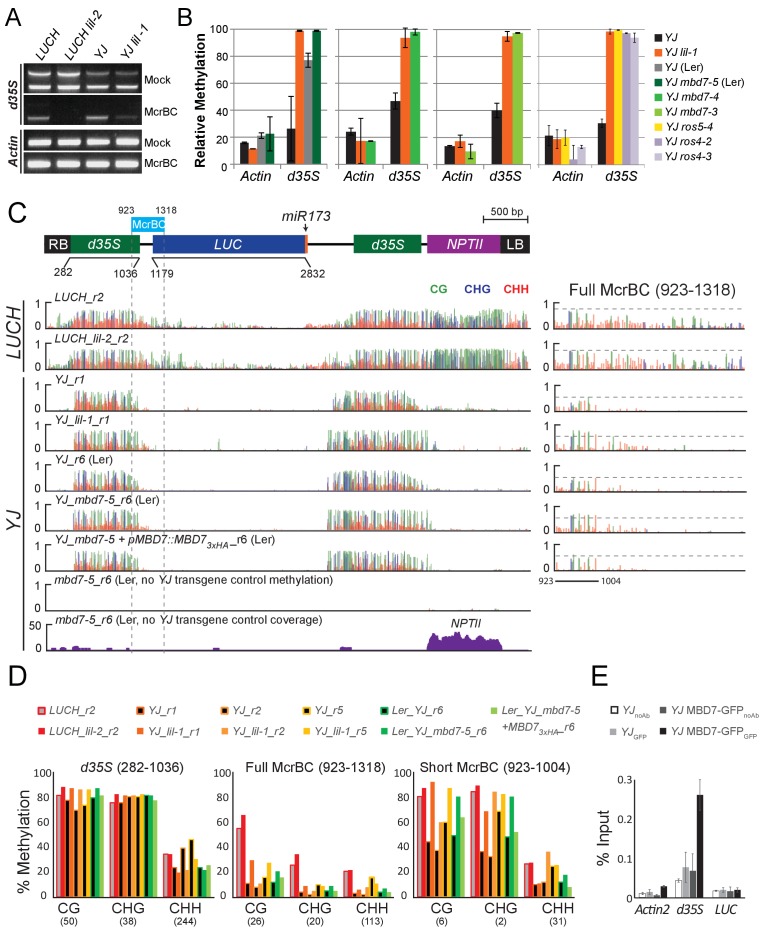
Figure 4. Disruption of the MBD7 complex results in subtle, but reproducible hyper-methylation at the d35S promoter.
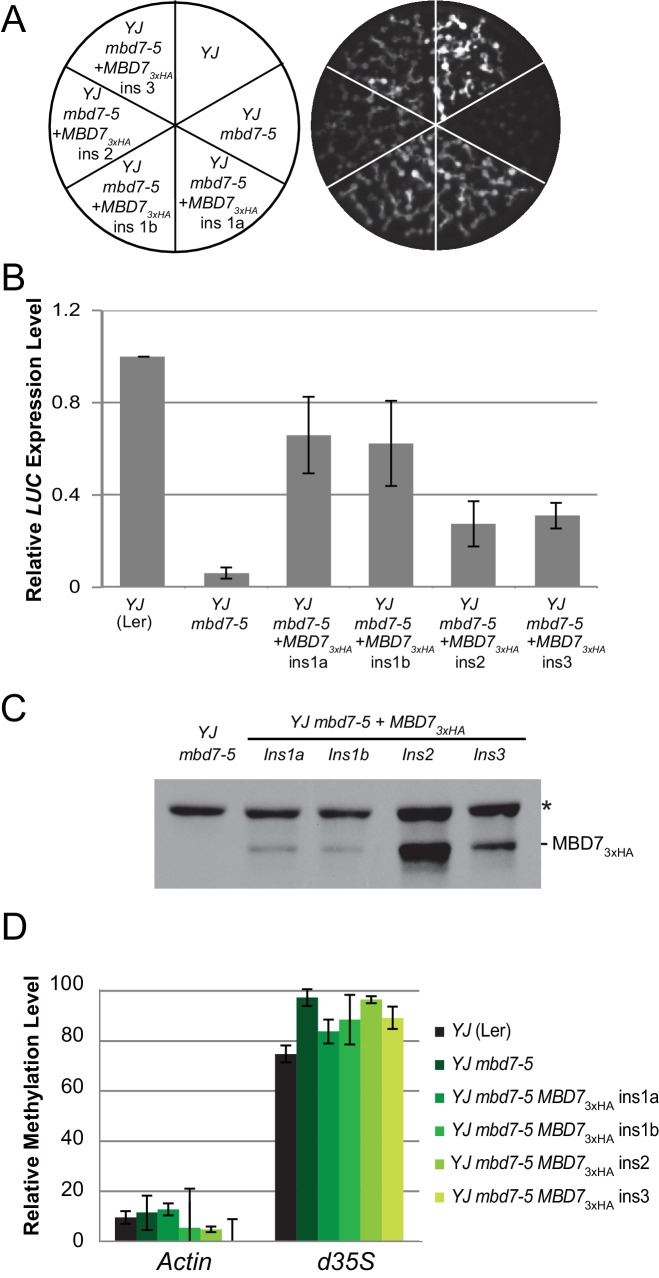
Cytosine methylation analysis by McrBC-PCR (A) and McrBC-qPCR (B) using mock or McrBC treated genomic DNA from the indicated genotypes. In (A), two tandem copies of the 35S sequence result in two PCR bands. ACTIN1, which lacks methylation, was used as the internal loading control. In (B), the relative methylation levels are plotted and the error bars indicate the standard deviation from two biological replicates. (C) A diagram of the YJ reporter drawn to scale relative to the browser tracks shown directly below. The light blue box indicates the region amplified for the McrBC assay. This region is expanded in the far right panel. DNA methylation tracks show the DNA methylation level (0–1, where 1 = 100% methylated) in the CG (green), CHG (blue), and CHH (red) sequence contexts at cytosines covered by at least five reads. The LUCH_r2 and YJ_r1 tracks correspond to those shown in Figure 1—figure supplement 1. The YJ mbd7-5 + pMBD7::MBD7-3xHA_r6 (Ler) line corresponds to the ‘ins1a’ line characterized in Figure 4—figure supplement 1. The coverage track for mbd7-5_r6 (bottom) shows the number of reads (y-axis) mapped to the transgene, demonstrating that sequence homology between the T-DNA in the mbd7-5 mutant and the LUC reporter is largely limited to the NPTII drug resistance gene. To facilitate visual assessment of the changes in DNA methylation, dashed horizontal lines are set relative to the maximal CG methylation at the 3’ end of the d35S promoter in the control LUCH and YJ reporters. (D) Quantification of the average percent methylation across the entire d35S promoter (282–1036), the Full McrBC region (923–1318) or a Short MrcBC region (923–1004) within the YJ transgene in the lines presented in (C). Each sample is appended with an ‘r#’ to indicate samples that were processed and sequenced together. Identical genotypes with different r#’s indicate biological replicates (e.g., YJ_r1, YJ_r2, and YJ_r5 are biological replicates). The number of cytosines in each sequence context within the quantified regions is indicated in parentheses. Note that the two d35S promoters driving LUC and NPTII are 94% identical in sequences, thus the DNA methylation data includes both multi-mapping and unique reads. (E) ChIP-qPCR showing enrichment of MBD7-GFP at the d35S promoter driving LUC expression at the YJ reporter. The data represents the average enrichment from three biological replicates as a percentage of the input and the error bars represent the standard deviation between replicates.