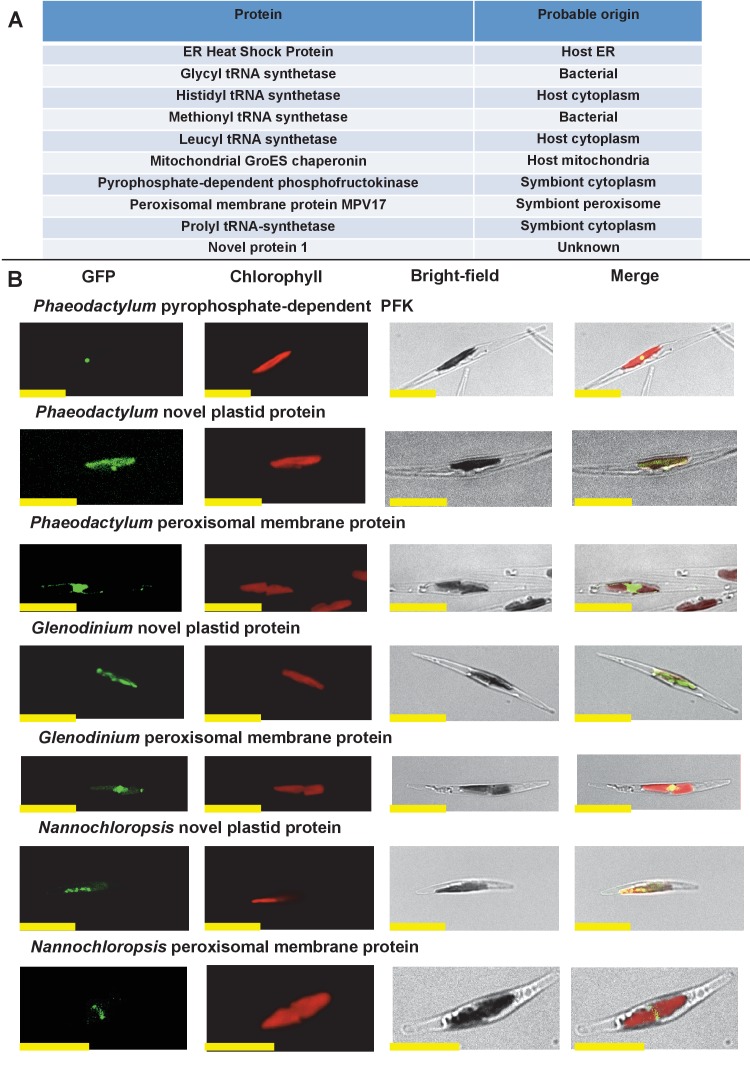
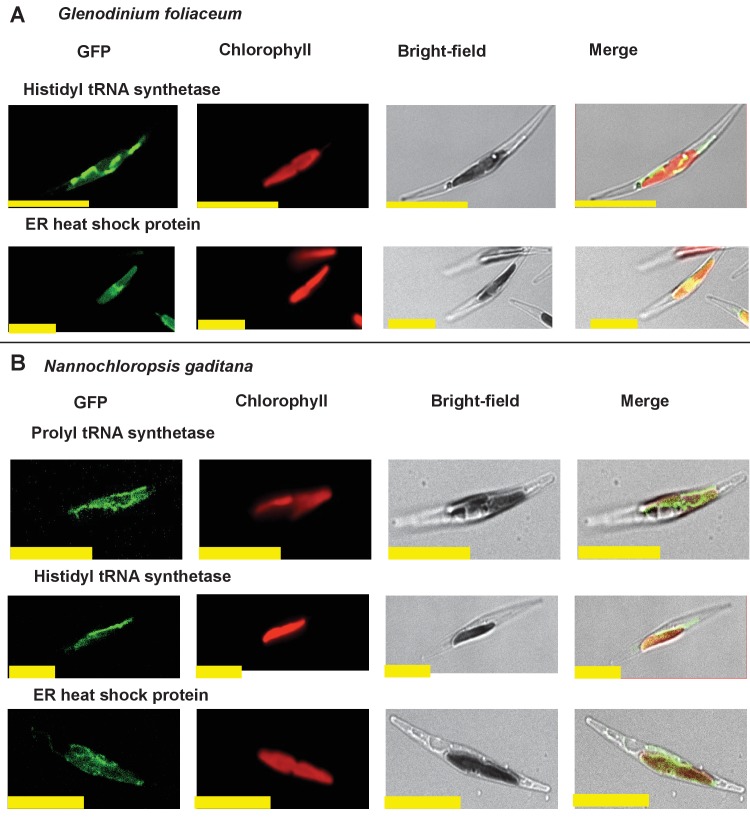
Figure 2. Verification of unusual ancestral plastid-targeted proteins.
(Panel A) lists the ten proteins selected for experimental characterisation and their most probable previous localisation prior to their establishment in the ochrophyte plastid, based on the first 50 nr BLAST hits. Exemplar alignments and single-gene tree topologies for some of these proteins are shown in Figure 2—figure supplements 1–4. (Panel B) shows the localisation of GFP constructs for copies of two proteins with an unambiguous plastid localisation (a pyrophosphate-dependent PFK, which localises to the pyrenoid, and a novel plastid protein, with cosmopolitan distribution across the plastid) and one protein with a periplastid localisation (a predicted peroxisomal membrane protein) from the diatom Phaeodactylum tricornutum, the diatom endosymbiont of the dinoflagellate Glenodinium foliaceum and the eustigmatophyte Nannochloropsis gaditana, expressed in P. tricornutum. All scale bars = 10 μm. Expression constructs for seven additional P. tricornutum proteins and three additional N. gaditana proteins with multipartite plastid localisations are shown in Figure 2—figure supplements 5 and 6, and control images (wild-type cells, and cells expressing untargeted eGFP) are shown in Figure 2—figure supplement 7.