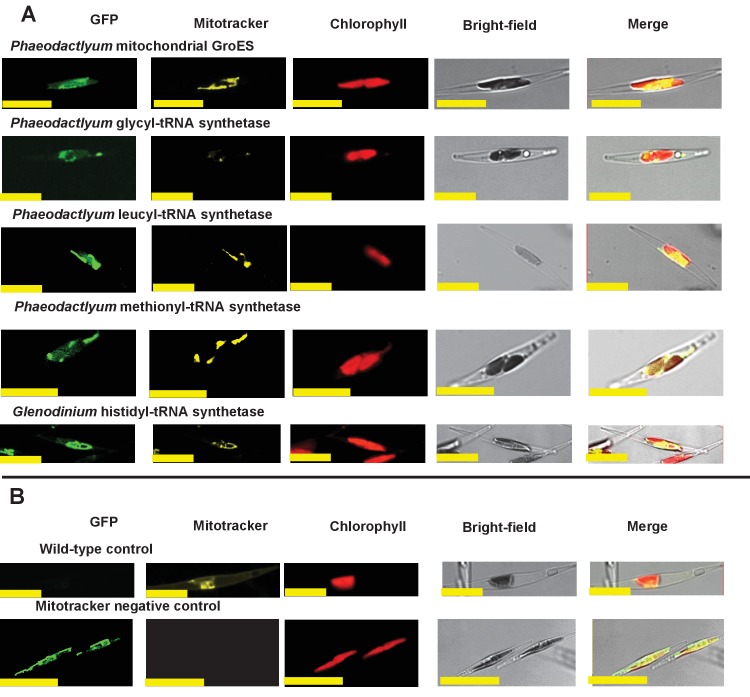
Figure 7. Ancient and bidirectional connections between the ochrophyte plastid and mitochondria.
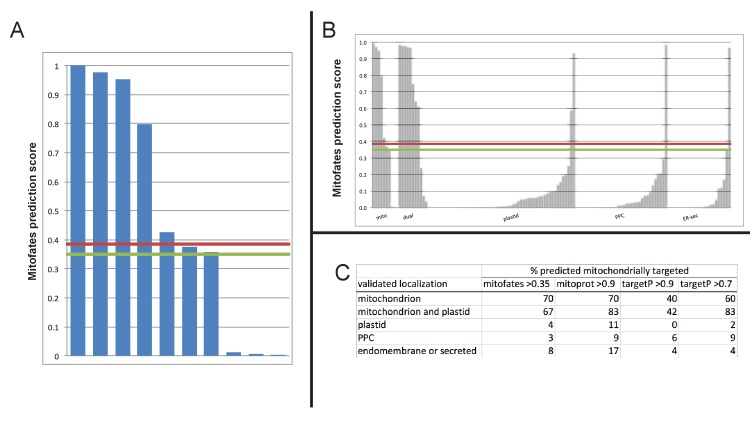
(Panel A) shows Mitotracker-Orange stained P. tricornutum lines expressing GFP fusion constructs for the N-terminal regions of histidyl- and prolyl-tRNA synthetase sequences from P. tricornutum and the eustigmatophyte Nannochloropsis gaditana. Targeting constructs for an additional four dual-targeted proteins in P. tricornutum and one dual-targeted protein in G. foliaceum, alongside Mitotracker-negative and wild type control images, are shown in Figure 7—figure supplement 1. (Panel B) profiles the predicted evolutionary origins of the 34 ancestral dual-targeted HPPGs, as inferred by BLAST top hit and single-gene tree analysis. Data supporting the thresholds used to identify probable dual-targeted HPPGs in silico are supplied in Figure 7—figure supplement 2. (Panel C) shows seven classes of tRNA synthetase for which only two copies were inferred in the genome of the last common ochrophyte ancestor. Evolutionary origins are inferred from combined BLAST top hit and single-gene tree analysis for dual-targeted proteins, and from BLAST top hit analysis alone for cytoplasmic proteins. In five cases the dual-targeted isoform is inferred to be of ultimate red algal origin, indicating that a protein derived from the endosymbiont has functionally replaced the endogenous host mitochondria-targeted copy.