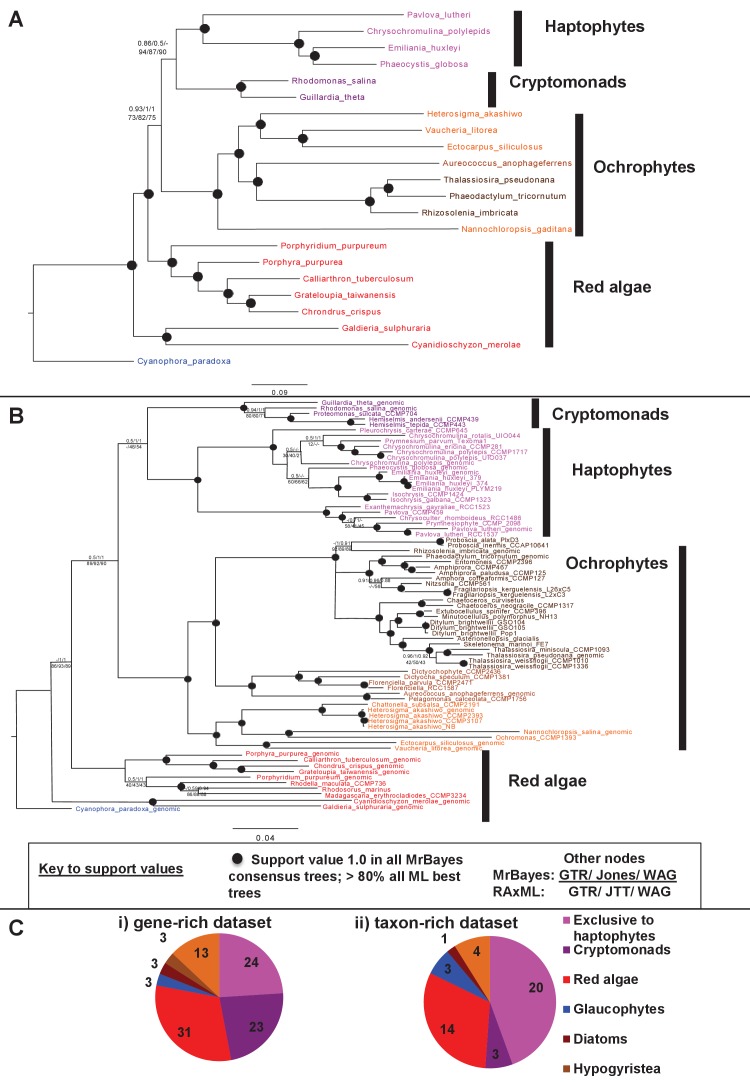
Figure 9. Non-ochrophyte origins of the haptophyte plastid genome.
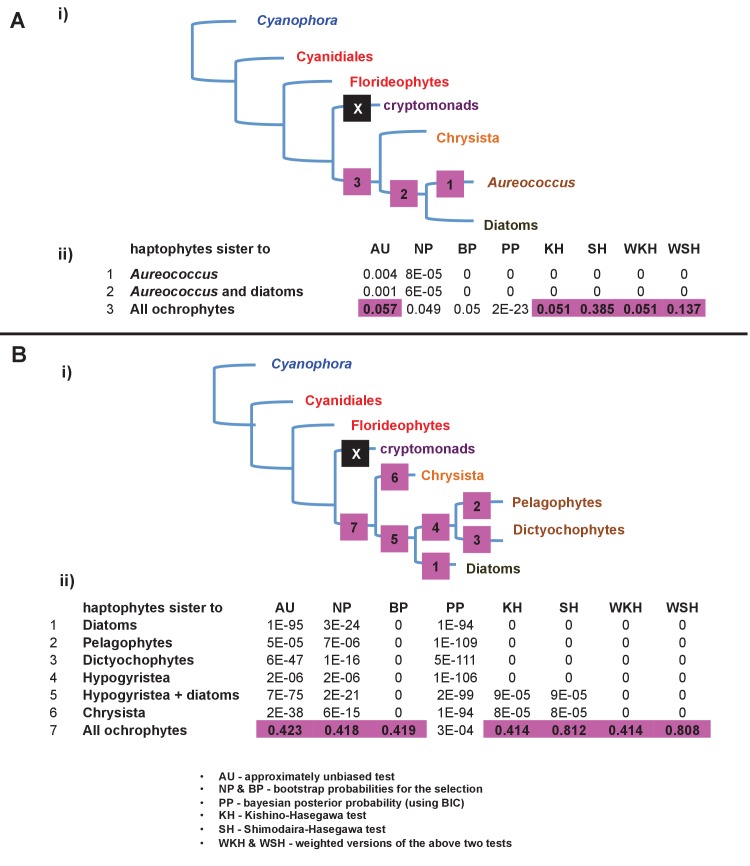
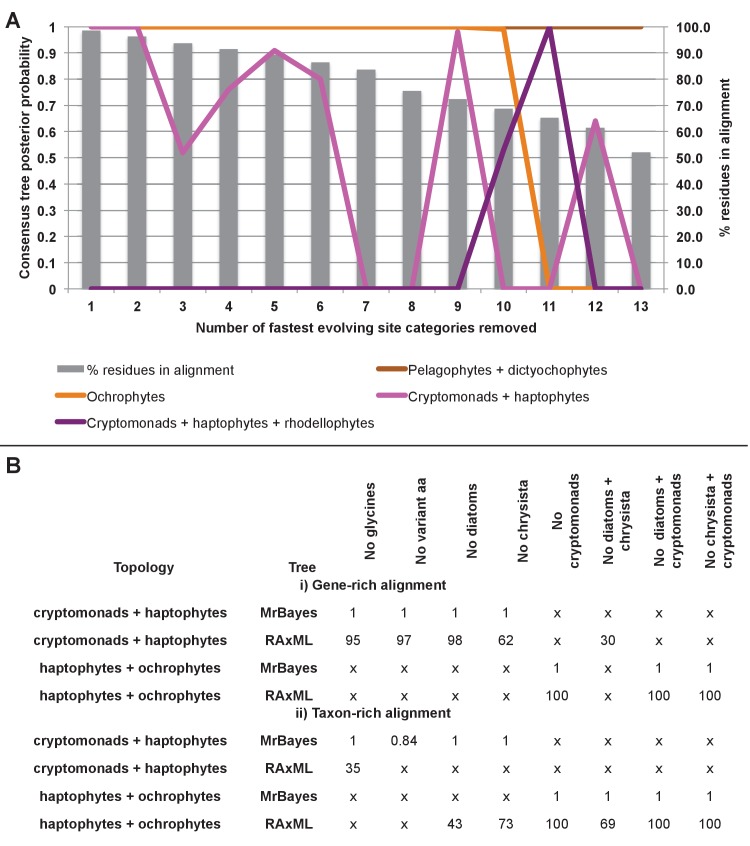
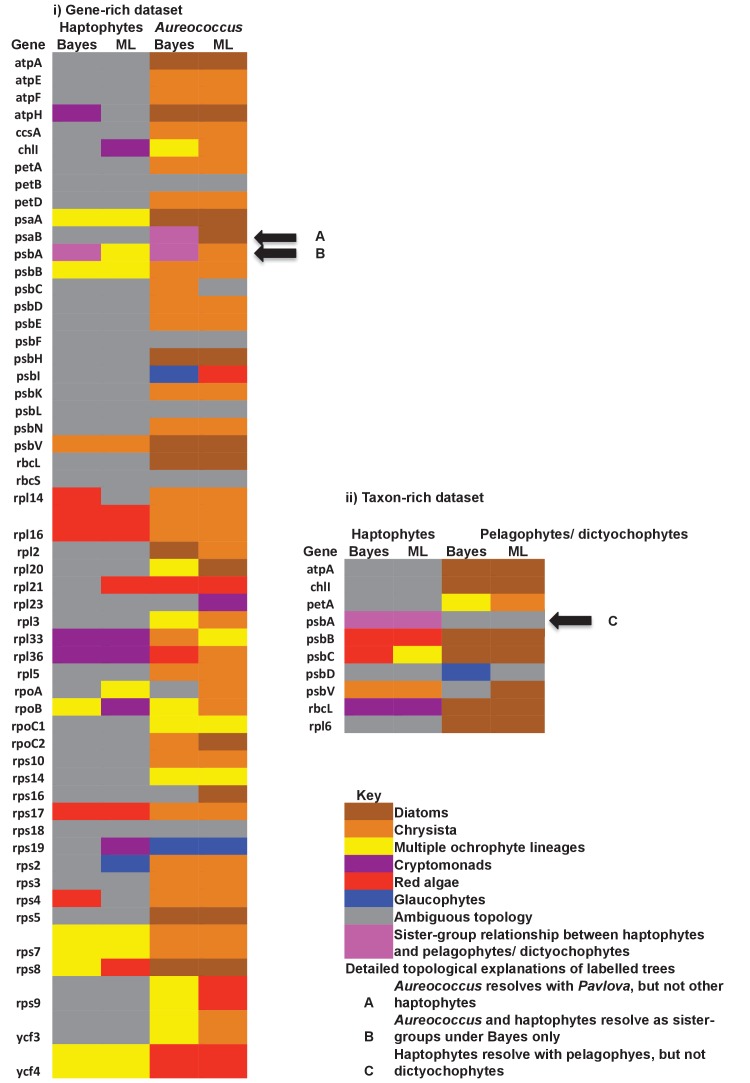
(Panels A and B), respectively, show gene-rich and taxon-rich phylogenies of plastid-encoded proteins from red algae and plastids of red algal origin with the glaucophyte Cyanophora paradoxa as outgroup. (Panel A) Combined Bayesian and Maximum Likelihood analysis (MrBayes + RAxML, GTR, JTT, WAG) of a 22 taxa x 12103 aa alignment of 54 proteins encoded by all published red and red-derived plastid genomes. (Panel B) analysis of a 75 taxa x 3737 aa alignment of 10 conserved plastid-encoded proteins detectable in a broad range of red lineage MMETSP libraries. Nodes resolve with robust support (posterior probabilities of 1 for all Bayesian trees and >80% bootstrap support for all ML trees) are shown with filled circles; individual support values for each analysis are shown for the remaining nodes are shown as detailed in the box below panel B. Alternative topology tests, the results of fast-site and clade deduction analysis for each tree, and heatmap comparisons of sister-group relationships identified for single-gene trees of each constituent gene within each concatenated alignment are shown in Figure 9—figure supplements 1–3. (Panel C) shows the number of residues in each alignment that are uniquely shared between haptophytes and only one other lineage. For the gene-rich alignment (i), which is gap-free, residues are included that are found in all four haptophyte sequences and at least one sequence from the lineage under consideration. For the taxon-rich alignment (ii), to account for the presence of gapped positions, residues are included that are found in at least 11 of the 22 haptophyte sequences and at least one sequence from the lineage under consideration.