Figure 5.
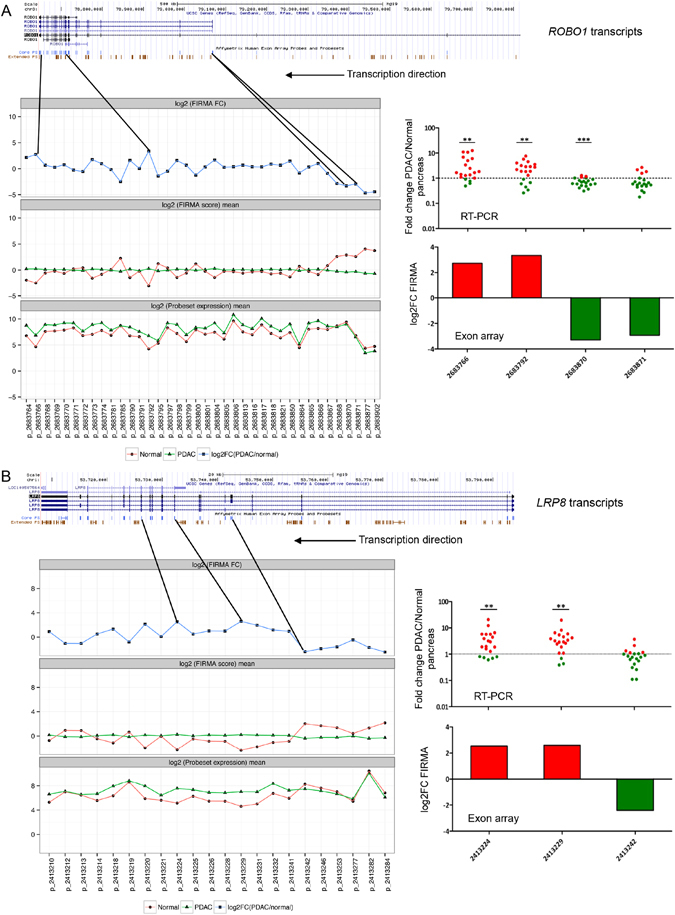
Analysis of multiple splicing events for ROBO1 and LRP8, and validation with RT-PCR. (A) Alternative splicing for ROBO1. (B) Alternative splicing for LRP8. The UCSC transcripts are shown at the top. For all quantified core probe-sets, means for probe-set expression and FIRMA values (log2 scaled), and the log2 FIRMA fold changes (PDAC/normal) were shown underneath UCSC transcripts. The probe-sets chosen for RT-PCR validation were shown with the black solid lines, with their positions within the transcripts indicated. RT-PCR results for the chosen probe-sets are represented as scatter dot plots of expression fold change in PDAC compared to normal pancreas, with the significance level shown at the top (* < 0.05, ** < 0.01, *** < 0.001). Corresponding exon array results are also reported in histograms of log2 FIRMA fold change (PDAC/normal) values. Both ROBO1 and LRP8 are coding for cell-surface proteins, and both were transcribed in the reverse strand.
