Summary
Immune cells function in diverse metabolic environments. Tissues with low glucose and high lactate concentrations, such as the intestinal tract or ischemic tissues, frequently require immune responses to be more pro-tolerant avoiding unwanted reactions against self-antigens or commensal bacteria. T-regulatory cells (Treg) maintain peripheral tolerance, but how Treg function in low glucose lactate rich environments is unknown. We report that the Treg transcription factor Foxp3 reprograms T cell metabolism by suppressing Myc and glycolysis, enhancing oxidative phosphorylation, and increasing nicotinamide adenine dinucleotide oxidation. These adaptations allow Treg a metabolic advantage in low glucose, lactate rich environments; resisting lactate mediated suppression of T cell function and proliferation. This metabolic phenotype may explain how Tregs promote peripheral immune tolerance during tissue injury, but also how cancer cells evade immune destruction in the tumor microenvironment. Understanding Treg metabolism may therefore lead to novel approaches for selective immune modulation in cancer and autoimmune diseases.
Keywords: T cell metabolism, immune regulation, immunometabolism
Introduction
T-regulatory cells (Treg) maintain peripheral tolerance and prevent autoimmune disease after tissue injury, when exposure of intracellular antigens not subject to central thymic selection may occur (Mueller, 2010; Warren and Strayer, 2013). The suppressive properties of Treg can also be harnessed by cancers, as tumors acquire mechanisms to evade host immunity (Nishikawa and Sakaguchi, 2010). Tumors arise from healthy tissue and many cancers acquire properties that enable growth independent from the host environment, including changes in cellular metabolism like aerobic glycolysis (Hanahan and Weinberg, 2011). The microenvironment surrounding solid tumors can be depleted of glucose, glutamine and tryptophan, while being enriched with lactic acid and kynurenines, thereby weakening anti-tumor immunity (Singer et al., 2011). This is an interesting parallel to tissues where immune tolerance is physiologically important, such as after ischemic injury or in the gastrointestinal tract.
Cytotoxic and effector T cells require glycolysis to proliferate and produce cytokines, and thus can become ineffective in glucose-depleted environments, which is a major problem for anti-tumor immunity (Chang et al., 2013; Chang et al., 2015; Ho et al., 2015; Macintyre et al., 2014). In contrast, Treg are less dependent on glycolysis and use oxidative phosphorylation (OXPHOS) for their energy production (Gerriets et al., 2015; Michalek et al., 2011), but the mechanisms explaining how Treg function in different metabolic environments remain relatively unexplored. Treg accumulate in the tumor microenvironment and are increasingly recognized as a therapeutic target in cancer immunotherapy (Nishikawa and Sakaguchi, 2014). We hypothesize that the known differences in energy metabolism between Treg versus cytotoxic and effector T cells may play a role in how Treg adapt to low glucose, lactate rich environments.
We have previously shown that Treg suppressive function can be augmented by targeting histone deacetylases (HDACs), which increases lysine acetylation of the key Treg transcription factor, Forkhead box P3 (Foxp3) (Beier et al., 2012; Tao et al., 2007). Acetylated Foxp3 shows resistance to proteasomal degradation, but also increased transcriptional activity (Liu et al., 2012; van Loosdregt et al., 2011). We have shown that deletion of HDAC9 increases OXPHOS in Treg (Beier et al., 2015), and found similar patterns in other HDAC deletion models, in which Foxp3 acetylation and Treg function were increased. This led us to hypothesize, that it may not just be a state in cellular metabolism such as OXPHOS that favors Treg differentiation and function (Lee et al., 2015), but also that Foxp3 exercises direct transcriptional control over T cell metabolism (Gerriets et al., 2016; Howie et al., 2017). Such Foxp3-mediated metabolic adaptations could enable Treg to maintain function in metabolically challenging conditions, where they can suppress inflammation as part of limiting injury, e.g. in ischemia (Brea et al., 2014), or, adversely, by impairing anti-cancer immunity in the tumor microenvironment (Nishikawa and Sakaguchi, 2010). Understanding how Foxp3+ Treg control their metabolism will have important therapeutic significance for anti-tumor immunotherapy, and aid in the development of new approaches to immunosuppression.
Results
Foxp3 expression induces OXPHOS
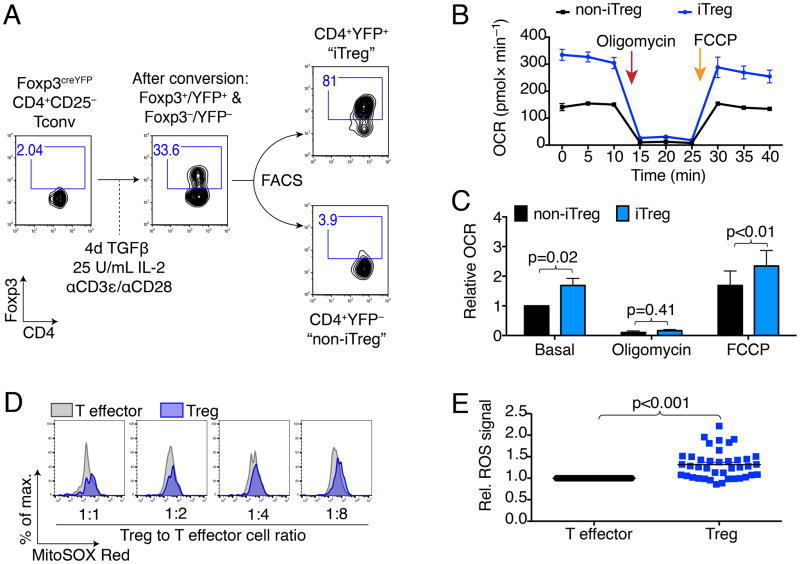
We started our investigation by examining if Foxp3 expression can reprogram T cell metabolism. We isolated CD4+CD25− conventional T cells (Tconv) from Foxp3YFPcre mice (Rubtsov et al., 2008), and cultured them under polarizing conditions to form iTregs. After 4 days, we sorted the cells into YFP+ ‘iTregs’ and YFP− ‘non-iTregs’ (Figure 1A & S1A). Using this approach, we could ensure that the stimulation conditions were identical; the cells were derived from the same culture wells, and differed only in their expression of Foxp3. Interestingly, Foxp3+ iTregs exhibited higher oxygen consumption rates (OCR) than Foxp3− non-iTreg (Figure 1B, C). Next, we examined thymic Treg (nTreg). In the absence of stimulation, nTreg have a very similar bioenergetics profile to Tconv cells (Figure S1B), which likely represents the absence of Myc expression described in resting T cells (Michalek et al., 2011; Wang et al., 2011). Two- and 16 h stimulated nTreg show an increase in OCR that is no different from stimulated Tconv, and, at 16 h is trending lower (Figure S1C–F). However, stimulated Tregs that are proliferating and not engaged in suppressive function can gradually lose their Treg phenotype (Hoffmann et al., 2009) or revert towards a glycolytic metabolism for proliferation (Gerriets et al., 2016). Co-culturing Tregs with effector T cells which stabilizes Foxp3 and the suppressive phenotyope (Akimova et al., 2011). To examine if the increase in iTreg OCR also correlated with improved OXPHOS in activated and phenotypically suppressive Treg with stable Foxp3 expression, we mixed Treg and Tconv cells at 1:1 to 1:8 ratios, and tracked reactive oxygen species (ROS) production by flow cytometry. In this approach, we used YFP to identify Foxp3YFPcre Treg, avoiding cell permeabilization for Foxp3 staining, which would have precluded ROS staining, and enabled us to distinguish the contribution of ROS from Tregs and effector T cells (Teff). Furthermore, we could ascertain that Treg maintain a suppressive phenotype by tracking suppression of the corresponding Teff. In these ‘working’ conditions, we observed that Treg had persistently elevated ROS production compared to Teff, independent of the Treg to effector T cell ratio and the degree of T effector cell suppression (Figure 1D, E). In a third model, we retrovirally transduced CD4+ T cells with MinR1-Foxp3 or empty vector constructs, as previously described (Liu et al., 2012). The retrovirally transduced T cells acquired suppressive function (Figure S1G) and showed a trend to increased OCR (Figure S1H, I), consistent with recent findings by the Rathmell group (Gerriets et al., 2016). We also questioned if Foxp3 acetylation could further influence T cell metabolism. However, using HDAC6 deletion and pharmacologic inhibition, Foxp3 acetylation specific effects could not be delineated from Foxp3 independent effects (Figure S2A–F). In conclusion, our data indicate that Foxp3 expression is sufficient to alter T cell metabolism and increase OXPHOS.
Figure 1. Expression of Foxp3 induces oxidative phosphorylation.
(A) Induced Treg (iTreg) model: Conventional T cells (CD4+CD25−, Tconv) were isolated from Foxp3YFPcre mice (Rubtsov et al., 2008), co-stimulated, and cultured under polarizing conditions to form iTreg, and then separated by fluorescence-activated cell sorting (FACS) into CD4+YPF+ “iTreg” and CD4+YFP− “non-iTreg”. Foxp3 and CD4 purity was assessed by flow cytometry. (B, C) Seahorse oxygen consumption rate (OCR) measurement using sorted iTreg and non-iTreg. (B) After obtaining basal respiration, the cells were subjected to 1.25 μM oligomycin, which inhibits ATP synthase and limits mitochondrial OCR. Subsequently, FCCP (cyanide-4-(trifluoromethoxy)phenylhydrazone) was added (0.5 μM), which uncouples mitochondrial respiration and maximizes OCR. (B) Representative OCR with iTreg exhibiting higher oxygen consumption than non-iTreg. (C) Pooled data from eight independent experiments (paired Student t-test), data shown after antimycin A correction. (D, E) Foxp3YFPcre Tregs and C57BL/6 T effector cells were stimulated with anti-CD3ε mAb and with irradiated CD90.2− antigen presenting cells. ROS production was measured after three days by superoxide sensitive fluorescence. Percentage of maximum (% of max) shows normalization of overlaid data and represents number of cells in each bin divided by the number of cells in the bin that contains the largest number of cells. (D) Foxp3YFP+ Treg had persistently increased ROS production than T effector cells from the same well. (E) Data pooled form three independent experiments with 40 observations, paired Student t-test. *, **, and *** indicates p<0.05, p<0.01, and p<0.001, respectively. Error bars indicate SEM.
Foxp3 suppresses Myc and glycolysis
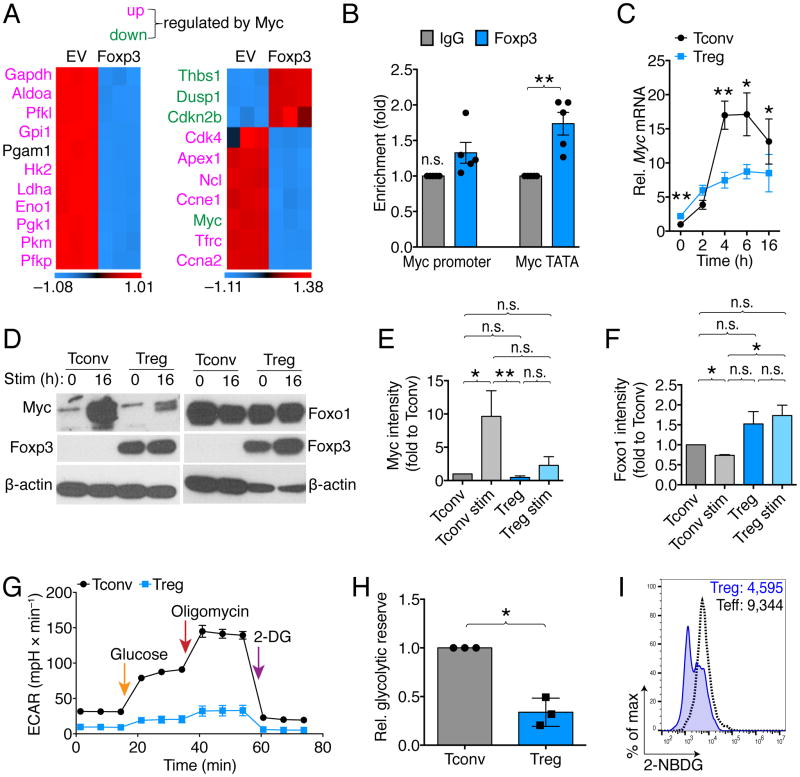
The observation that Foxp3 expression is sufficient to increase OCR in T cells suggests that Foxp3 might transcriptionally control cellular metabolism. To screen for potential Foxp3 targets, we performed ChIP-chip of HDAC6KO and WT Tregs, precipitating genomic DNA segments with Foxp3 mAb. We have previously shown that HDAC6KO Tregs have more acetylated and transcriptionally active Foxp3 (Beier et al., 2012). In addition to expected Foxp3 transcriptional targets known to be increased in HDAC6KO with more acetylated Foxp3, such as Ctla4 (de Zoeten et al., 2011), we found Foxp3 binding at the Myc promoter (Figure S2G). Myc is a transcription factor, which has important roles in upregulating glycolysis and glutaminolysis in activated T cells (Wang et al., 2011). Since the iTreg model requires TGF-β for cell culture, which itself can influence T cell activation and metabolism, we characterized the interaction of Myc and Foxp3 in models independent of TGF-β. First, we analyzed microarray gene expression data from a study in which CD4+ T cells underwent retroviral transduction with Foxp3 or empty vector control (Liu et al., 2012). T cells transduced with Foxp3 by this method acquire a suppressive phenotype similar to Treg (Figure S1G). We analyzed Foxp3-dependent gene expression by assessing genes that are known to be Myc-dependent (Zeller et al., 2003), and found that both glycolytic Myc target genes, as well as non-glycolytic Myc targets, were strongly downregulated when Foxp3 was present (Figure 2A). Outside of T cells, Foxp3 has been reported as a tumor suppressor and Myc inhibitor in glioblastoma cells (Frattini et al., 2012). To assess if Foxp3 can bind to the Myc promoter in Treg cells, we performed ChIP assays in resting and 16 h CD3ε/CD28 co-stimulated Tregs, as well as in vivo expanded Treg cells, and found increased Foxp3 binding to the Myc TATA box (Figure 2B). Next, we assessed Myc gene expression upon activation of Treg and Tconv cells. As expected, Tconv cells strongly upregulated Myc mRNA upon CD3ε/CD28 co-stimulation, especially between 4–6 hours after stimulation, while in Treg Myc mRNA was not induced (Figure 2C). The mRNA results corresponded with protein data, which consistently showed that Tregs do not upregulate Myc after 16 h stimulation (Figure 2D, E). Another candidate for Treg metabolic control includes Foxo1, which is stabilized in Treg by less protein kinase B (Akt) signaling and increased activity of phosphatase and tensin homolog, PTEN (Huynh et al., 2015; Ouyang et al., 2010). We also found Foxo1 better preserved in Treg after activation, relative to Tconv (Figure 2D, F). Foxo1 is well described to suppress glycolysis and increase lipid oxidation (Gross et al., 2008). Consistent with the Myc and Foxo1 data, unlike Tconv, Treg did not adopt a strong glycolytic response when exposed glucose or oligomycin (Figure 2G, H). In addition, Treg exhibited reduced glucose uptake (Figure 2I), consistent with recent reports on Foxp3-dependent suppression of Glut1 (Gerriets et al., 2016). Beyond Myc and Foxo1, we hypothesized additional transcriptional control of Foxp3 over nuclear encoded genes controlling the electron transport chain (ETC) in mitochondria. A previous study investigating the transcriptional profile of Foxp3 identified Foxp3 binding to Ndufv2, which encodes NADH dehydrogenase flavoprotein 2 and is part of ETC complex I (Marson et al., 2007). We found an increase of ETC complex I in iTreg relative to FACS-separated non-iTreg, but did not see it in nTreg (Figure S3A–C). Ndufv2 mRNA was not significantly different between Tconv and Treg, and neither was Uncoupling protein-2 mRNA expression (Figure S2D, E). In summary, Foxp3 expression decreases gene expression of Myc-dependent transcripts, and Foxp3 binds to the Myc TATA box. Foxp3+ Treg do not upregulate Myc upon CD3ε/CD28 stimulation, and have only a limited response to stimuli that activate glycolysis.
Figure 2. Foxp3 suppresses Myc and glycolysis.
(A) Microarray analysis showed suppression of Myc signaling in Foxp3 vs. empty vector (EV) transduced T cells (3/group). (B) Chromatin immunoprecipitation of gDNA from 16 h co-stimulated Treg using Foxp3 pulldown showed identifies the TATA box of Myc as a Foxp3 binding site. Data pooled from five independent experiments (paired Student t-test). (C) Tconv and Treg were co-stimulated and cultured with 25 U × ml−1 IL-2, and RNA was obtained at the indicated time points. Tconv strongly upregulated Myc mRNA while Treg did not. Data pooled from six independent experiments (paired Student t-test). (D, E) Western blotting shows that unlike Tconv, co-stimulated Treg are unable to upregulate Myc, and showed better preservation of Foxo1. (D) Representative and (E, F) pooled data from six (Myc) and three (Foxo1) independent experiments, normalized to freshly isolated Tconv (paired one-way ANOVA). (G) Extracellular acidification rate (ECAR) measurements of 16 h co-stimulated Treg and Tconv indicates diminished capacity of Tregs to mount a glycolytic response. (G) Pooled glycolytic reserve data related to (F), 3/group, paired Student t-test. (H) Treg and Tconv were stimulated with soluble anti-CD3ε mAb and irradiated antigen presenting cells. After three days, effector T cells took up more fluorescent-labeled glucose (2-NBDG) than Treg. Data representative of two independent experiments. * and ** indicate p-values <0.05 and <0.01, respectively. Error bars indicate SEM. Abbreviations: Aldoa, Aldolase A; Pfkl, 6-phosphofructokinase, liver; Gpi, Glucose-6-phosphate isomerase; Pgam, Phosphoglycerate mutase; Hk, Hexokinase; Ldha, Lactate dehydrogenase A; Eno, Enolase; n.s., not significant; Pgk, Phosphoglycerate kinase; Pkm, pyruvate kinase, muscle; Pfkp, Phosphofructokinase, platelet; Thbs, Thrombospondin; Dusp, Dual specificity protein phosphatase; Cdkn2b, Cyclin-dependent kinase 4 inhibitor B; Cdk, Cyclin-dependent kinase; Apex, DNA-(apurinic or apyrimidinic site) lyase; Ncl, Nucleolin; Ccne1, Cyclin E1; Trfc, Transferrin receptor 1; Ccna2, Cyclin A1; 2-DG, 2-Deoxy-D-glucose.
ETC complex I, but not complex IV, is essential for Treg function
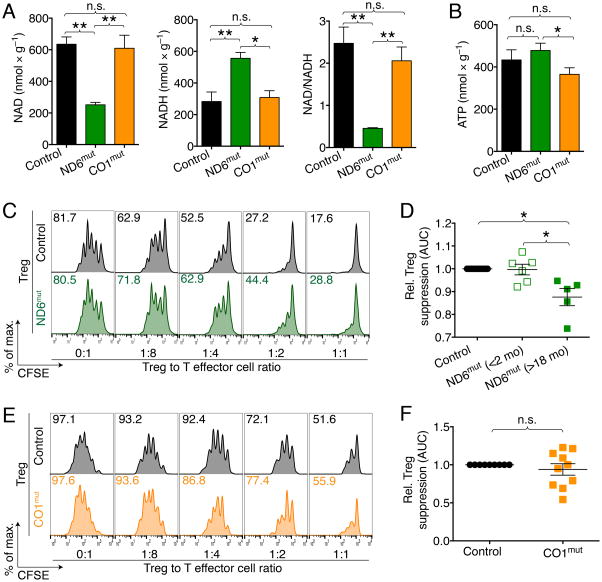
Given the importance of the mitochondrial respiratory chain for OXPHOS, and the finding of increased OCR after Foxp3 expression, we questioned if different complexes and functions of the electron transport chain (ETC) were important to Treg function. To that end, we utilized two different mouse models of mitochondrial defects. The first model has impaired ETC complex I, where the mitochondrial ND6 gene has a mutation, mt-ND6P25L, hereafter referred to as ND6mut (Lin et al., 2012). In the second model, we used mice with a mtDNA CO1 mutation (CO1V421A, CO1mut hereafter), which have a 50% reduction in ETC complex IV. The two models are distinct by the selective impairment in either ETC complex I or complex IV. ND6mut mice show impaired oxidation of nicotinamide adenine dinucleotide hydrate (NADH) to nicotinamide adenine dinucleotide (NAD), while having normal ATP production. In contrast, CO1mut mice have normal NADH to NAD oxidation, but show impaired ATP production (Figure 3A, B & S4A, B). T cell development was not impaired in either model (Figure S4C–H). We found that Tregs from ND6mut, but not CO1mut mice, had reduced suppressive function (Figure 4C–F). Similar effects to the genetic targeting of the NADH dehydrogenase complex can be achieved with the complex I inhibitor, rotenone (Beier et al., 2015; Gerriets et al., 2015). In summary, ETC complex I, but not complex IV, is important to Treg function. This suggests that ETC complex I specific functions such as oxidation of NADH to NAD may be of particular relevance to Treg function.
Figure 3. Impairment in ETC complex I, but not complex IV, reduce Treg suppressive function.
(A) NAD/NADH and (B) ATP measurements in heart tissue from age and gender matched ND6mut (ETC complex I defect), CO1mut (ETC complex IV defect) and co-housed wild type control mice (4–6/group). ND6mut mice show reduced NAD relative to NADH, with normal ATP production. CO1mut mice are capable of NADH to NAD oxidation, but show impaired ATP production. (C, D) Comparison of the ability of ND6mut versus control Tregs to suppress proliferation of CFSE-labeled T-effector (Teff) cells in vitro, stimulated with anti-CD3ε mAb and irradiated CD90.2− antigen presenting cells. (C) Representative flow cytometry and (D) cumulative analysis showing Treg from aged ND6mut mice have weaker suppressive Treg function versus results for WT Treg. Student’s t-test; data pooled from 11 observations and four independent experiments. (E) Comparison of the ability of CO1mut versus control Tregs shows no difference in suppressive function. (F) Cumulative data; Student’s t-test. Data pooled from 10 observations and three independent experiments. All wild type controls were age- and gender matched. * indicates p<0.05, and error bars indicate SEM.
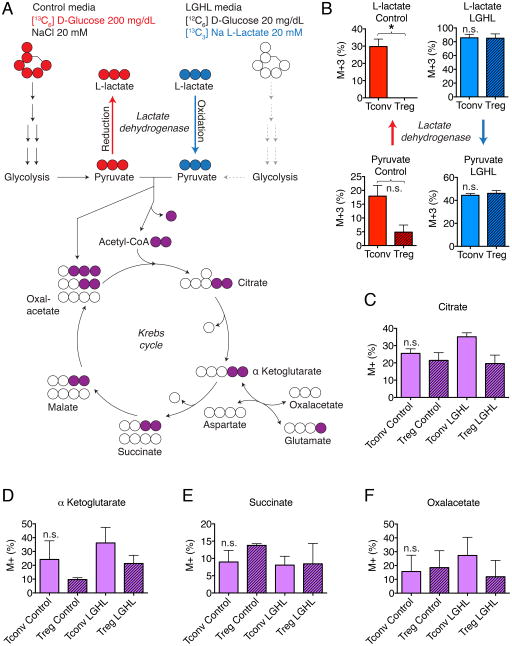
Figure 4. Tregs favor oxidation of L-lactate to pyruvate over reduction of pyruvate to L-lactate.
(A) Experimental design. (B) Left panels (red): Heavy glucose derivative analysis of M+3 pyruvate and M+3 L-lactate (with 3 × [13C]-atoms and no [12C]-atoms) shows that Tconv form more pyruvate and L-lactate than Treg. M+3 L-lactate was undetectable in Tregs. Right panel (blue): Both Tconv and Treg convert L-lactate to pyruvate. (C-F) Tricarboxylic acid derivative analysis shows that both Treg and Tconv integrate glucose and L-lactate into the Krebs cycle. Data pooled from three independent experiments (paired Student t-test). Abbreviations: LGHL, Low glucose, high L-lactate. M+ refers to tricarboxylic acid derivatives with at least one [13C]-atom (after background correction). Error bars indicate SEM.
Foxp3 expression limits pyruvate to lactate reduction by Lactate dehydrogenase
Our findings raised the question if Foxp3 expression can redirect the metabolic behavior of T cells and adapt them to different environments. We utilized ultra-high-performance liquid chromatography-selected reaction monitoring/mass spectrometry to track the derivatives of L-lactate and D-glucose in 16 hour stimulated Treg and Tconv (Figure 4A & S5A, B). We exposed both Tregs and Tconv to two types of media. In one condition, we used low glucose high L-lactate (LGHL, 20 mg × dl−1 [12C6]-D-glucose, 20 mM Na [13C3]-L-lactate). In the control media, we used 200 mg × dl−1 [13C6]-D-glucose with added 20 mM NaCl. The control media with stable isotope labeled [13C6]-glucose allowed us to track glucose utilization and tracking of glucose metabolites, while the LGHL media allowed following [13C3]-L-lactate derivatives. As expected based upon our studies on the Foxp3-Myc interaction and glycolysis, we found that Treg produced less [13C3]-pyruvate from [13C6]-D-glucose. Interestingly, activated Tconv produced a significant amount of [13C3]-L-lactate, which was undetectable in Tregs in each of three independent experiments (Figure 4B & S5C). This is remarkable, because the M+3 of pyruvate in the Tconv was 17.9% (±3.9%) of the pyruvate pool and this increased to 29.8% (±4.3%) in the L-lactate pool. These findings revealed that a significant proportion of glucose in Tconv is metabolized to pyruvate, which in turn is reduced by lactate dehydrogenase (LDH) to L-lactate. However, in Tregs, this does not happen. Both Tconv and Treg can oxidize L-lactate to pyruvate (Figure 4B), and both form tricarboxylic acid derivatives (Figure 4C–F & S5D). Both effector T cells and Treg have near equal quantities of tricarboxylic acid derivatives. Retroviral transduction of Foxp3 into T cells (vs. empty vector control) shows a slightly increased [13C]-enrichment of most Krebs cycle intermediates in Foxp3-transduced T cells (Figure S5E). In summary, the lack of L-lactate production by Treg suggest that Foxp3 can influence the direction of the LDH reaction favoring oxidation of L-lactate to pyruvate while in LGHL conditions, both Treg and Tconv oxidize L-lactate to pyruvate.
L-lactate impairs effector, but not regulatory T cell function
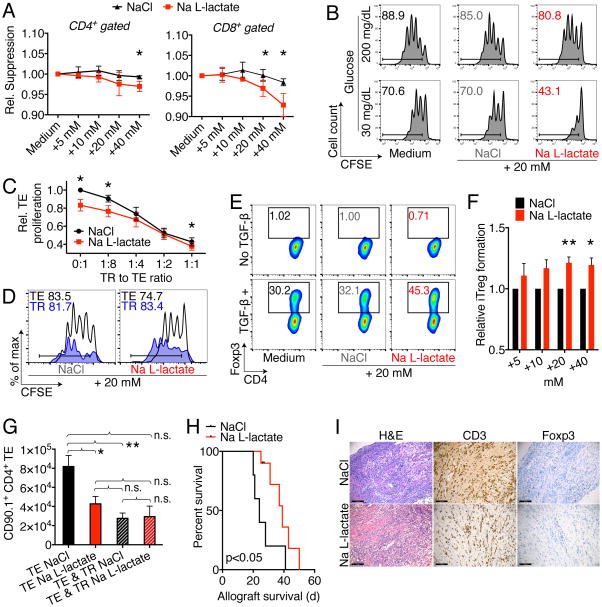
Based upon our findings that Foxp3 regulates T cell metabolism by suppressing Myc, the inability of Treg to form L-lactate, and the effective turnover of L-lactate to pyruvate in Treg cells, we hypothesized that Treg cells may be able to withstand the strong suppressive effects that L-lactate has on effector and cytotoxic T cells (Calcinotto et al., 2012; Fischer et al., 2007). We found that proliferating murine (and human) T cells exposed to increasing levels of Na L-lactate showed suppression of proliferation, independent of acidity and toxicity (Figure 5A, B & S6A). In contrast, Treg suppressive function was unaffected by the addition of Na L-lactate, just like they were resistant to the suppressive effects of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) inhibition (Figure 5C & S6B). While Teff proliferation was reduced, Treg division was unimpaired (Figure 5D). We noted that Na L-lactate added to Tconv cells, cultured under polarizing conditions to form iTreg, led to an increase in iTreg formation (Figure 5E, F & S6C). Together, our findings support an immunosuppressive role of L-lactate, suppressing CD8+ and CD4+ T cells, but sparing (and in fact inducing) Tregs. Added Na L-lactate did not influence T cell metabolism or ROS dependent stress signaling (Figure S6D-F). Na L-lactate did increase Th17 polarization (Figure S6G, H). Next, we tested if the observed immunosuppressive effects of L-lactate can be seen in vivo. We observed significant reduction of Teff homeostatic proliferation, as well as a short prolongation of MHC-mismatched, BALB/c to C57BL/6 cardiac allograft survival (Figure 5G–I, S6I, J). Together, these data show that L-lactate can impair T cell proliferation, but that Treg proliferation and function is not negatively affected, and iTreg development even favored by L-lactate.
Figure 5. L-lactate impairs T effector cells but not Tregs.
(A) C57BL/6 splenocytes were labeled with CFSE and stimulated with 1 μg × ml−1 soluble CD3ε mAb, and CD4+ as well as CD8+ T cell proliferation was assessed after three days. Incremental doses of Na L-lactate suppressed T cell proliferation compared to NaCl control. Data pooled from four independent experiments (paired Student t-test). (B) CD4+CD25− Tconv were CFSE labeled and stimulated with anti-CD3ε mAb and irradiated antigen presenting cells. Reduced ambient glucose augmented the suppressive effect of pH neutral Na L-lactate suppression of T cell proliferation. Data representative of three independent experiments. (C) CFSE labeled effector T cells (TE) were co-stimulated with anti-CD3ε mAb and irradiated CD90.2− antigen presenting cells, and suppressed by adding regulatory T cells (TR) at the indicated ratios. In contrast to TE proliferation, TR suppressive function is not impaired by added Na L-lactate (7/group, paired Student t-test). (D) Treg suppression assay with CD90.1 Treg and CD90.2 effector T cells to track proliferation of each subpopulation. Na L-lactate impaired Teff proliferation, but not TR. Data representative of three independent experiments. (E) Representative and (F) quantified data from 10 independent Treg induction experiments under the same conditions as in Fig 1A (paired Student t-test). Added pH neutral Na L-lactate augments induced Treg formation. (G) B6/Rag1−/− mice were adoptively transferred with 1 × 106 Tconv ± 2.5 × 105 Treg cells, and treated i.p. with 60 μl × g−1 of 150 mM Na L-lactate or NaCl control for seven days. Administration of Na L-lactate reduced TE (CD90.1+CD4+) homeostatic proliferation without affecting TR suppressive function. * and ** indicates p<0.05 and p<0.01, respectively (3–11/group, ANOVA with Tukey’s multiple comparisons test). (H, I): Cardiac allografts from BALB/c (H-2d) donors were transplanted into the abdomens of MHC-mismatched C57BL/6 (H-2b) recipients. Recipients were then treated for 14 days with 0.2 mg × kg−1 × d−1 rapamycin i.p., and 60 μl × g−1 body weight of 150 mM Na L-lactate (9 μmol × g−1 × d−1, 10/group) or NaCl (5/group) control. (H) Na L-lactate injections led to a brief prolongation of allograft survival (Mantel Cox test). (I) Allograft histology at 20–28 d post-transplant shows a similar degree of lymphocyte infiltration, with equal CD3 and Foxp3 cells (4–5/group). Error bars indicate SEM.
The effect of L-lactate on T cell proliferation and function is LDH dependent
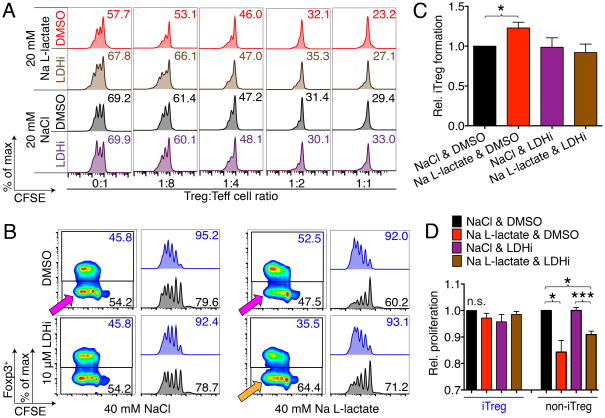
The finding that Foxp3 adjusted T cell metabolism and conveyed resistance to the suppressive effects of L-lactate led us to the hypothesis the L-lactate mediated suppression is LDH-dependent. We found that C1 esters of L-lactate, which are resistant to LDH oxidation, do not mediate Teff suppression in vivo (Figure S7A–C). To assess the role of LDH directly, we combined L-lactate T cell function studies with an LDH inhibitor (LDHi), GSK 2837808A (Billiard et al., 2013; Xie et al., 2014). As expected, Na L-lactate treatment reduced Teff proliferation without affecting Treg function (Figure 6A, S7D–F). Interestingly, we found that LDHi treatment did not impair Treg suppressive function, but rather, it rescued the Teff from L-lactate inhibition. We made a similar observation in our iTreg model (Figure 6B–D, S7G, H). Here, we CFSE labeled Tconv cells and polarized them to iTreg under low glucose conditions (20 mg/dl). We found that only the T cells that acquired Foxp3 were able to withstand the suppressive L-lactate, while the ‘non-iTreg’ were impaired in their ability to proliferate (Figure 6B, pink arrows). We had originally expected to find that LDH inhibition would negate this ‘metabolic advantage’ and deny Foxp3+ iTreg the ability to use L-lactate as alternative fuel source when glucose is sparse; however, instead we found that LDHi rescued the non-iTreg from impaired proliferation (Figure 6B, orange arrow, and 6D), as it did the Teff cells (Figure 6A). These data suggested that the Treg ‘metabolic advantage’ does not depend upon the ability to feed on L-lactate as an alternative fuel, but rather, that the impairment of Teff function depends on the oxidation of L-lactate to pyruvate by LDH.
Figure 6. Effects of L-lactate on T cell function are LDH dependent.
(A) CFSE labeled effector T cells (Teff) were co-stimulated with anti-CD3ε mAb and irradiated CD90.2− antigen presenting cells in low glucose media (20 mg × dl−1), and suppressed by adding regulatory T cells (Treg) at the indicated ratios. Teff proliferation was impaired with Na L-lactate, and restored by addition of the LDH inhibitor (LDHi) GSK 2837808A. Data representative of three independent experiments. (B-D): CD4+CD25− Tconv were CFSE labeled and cultured under polarizing conditions to form iTreg (see Fig. 1A) in low glucose media (20 mg × dl−1). After 4 days, Foxp3+ iTreg formation was assessed. Na L-lactate did, as expected (Fig. 6E, F) increase iTreg formation. This was reverted by LDH inhibition. (B) Quantitative (4/group) and (C) representative data. (C) Interestingly, iTreg that upregulated Foxp3 were not impaired in their ability to proliferate, whereas T cells that did not upregulate Foxp3 (‘non-iTreg’) were suppressed by the addition of L-lactate (pink arrows). Exposure to a LDH inhibitor reversed this suppression, and protected the non-iTreg from impaired proliferation by Na L-lactate. (D) Quantitative proliferation data, 4/group. *, **, and *** indicates p<0.05, p<0.01, and p<0.001, respectively (paired one-way ANOVA for B & D). Error bars indicate SEM.
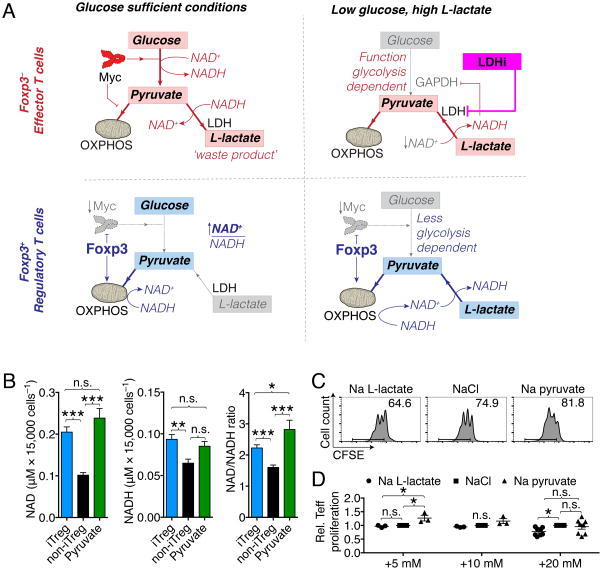
Foxp3 expression increases the NAD:NADH ratio
The observation that Teff and non-iTreg proliferation was suppressed by L-lactate was further supported by administering D-lactate, which is not metabolized through LDH (EC 1.1.1.27), but through the D-lactate specific LDHD (EC 1.1.1.28); LDHD also reduces NAD to NADH upon D-lactate to pyruvate oxidation (Ewaschuk et al., 2005). We observed that LDHD function is not affected by GSK 2837808A (Figure S7F). Interestingly, the suppressive effects of D-lactate on non-iTreg can be rescued only to a lesser degree by LDH inhibition (Figure S7G), further supporting the observation that the inhibitory effects of lactate on T cell proliferation are LDH dependent. These observations led us to the hypothesis that the ‘metabolic advantage’ of Treg could be based on resistance to NAD depletion (Figure 7A). We propose that Foxp3 is responsible for shutting off glycolysis through Myc and potentially through other mechanisms like Foxo1 and PTEN (Gerriets et al., 2016; Huynh et al., 2015; Ouyang et al., 2010). Suppression of Myc also increases pyruvate dehydrogenase activity (Dang et al., 2008). Thus, Foxp3+ Treg have increased NAD production (OXPHOS) and reduced NAD consumption (glycolysis). In contrast, cytotoxic and effector T cells reduce NAD to NADH via GAPDH, and require constant NAD recycling through LDH, producing L-lactate in the process (Figure 7A). Indeed, we found that iTreg have significantly increased NAD (Figure 7B & S7I). Furthermore, in contrast to the effects of Na L-lactate, Teff proliferation in low glucose environments can be augmented by adding Na pyruvate, which increases the NAD:NADH ratio (Figure 7C, D & S7I).
Figure 7. Foxp+ Treg have higher NAD:NADH ratios.
(A) Conceptual model of how Foxp3+ Treg can escape the suppressive effects of low glucose, high L-lactate environments. (B) CD4+CD25− Tconv were isolated from Foxp3creYFP mice, co-stimulated, and cultured under polarizing conditions to form iTreg, and then separated by fluorescence-activated cell sorting (FACS) into CD4+YPF+ “iTreg” and CD4+YFP− “non-iTreg” (see Figure 1A). After separation, the cells were re-exposed to stimulation conditions for one hour, and then harvested for NAD and NADH measurements. Expression of Foxp3 increased the fraction of NAD. Errors bars indicate SEM. (C, D) Tconv were CFSE labeled and co-stimulated for three days with soluble CD3ε mAb and irradiated antigen presenting cells in low glucose media (20 mg × dl−1) with added 5–20 mM of either NaCl, Na L-lactate, or Na pyruvate. (C) Flow cytometry (+20 mM) and (D) pooled data from 3–8 independent experiments shown. * indicates p<0.05 (one-way ANOVA), error bars indicate SEM.
Discussion
Immune tolerance relies on thymic selection as well as peripheral mechanisms preventing sensitization to self-antigens (Mueller, 2010). Tissue injury risks exposure of antigens such as histones and double stranded DNA that exceed the clearance capacity of phagocytic cells, which may predispose to autoimmune disease (Mok and Lau, 2003). Our data show that Treg cells exhibit adaptations that enable a selective metabolic advantage in low glucose, high lactate environments. Teff require NAD regeneration to maintain GAPDH activity and glycolysis. At 20 mg × dl−1 glucose, T cells still have enough glucose to fuel proliferation, yet the influx of L-lactate and the reversal of the LDH reaction shuts down their ability to regenerate NAD, scuttling glycolysis, while LDH inhibitor treatment restored the ability of Teff to proliferate. Loss of glycolysis is particularity detrimental for effector and cytotoxic T cells, as it impairs the ability to produce IFN-γ or maintain intracellular calcium (Chang et al., 2013; Chang et al., 2015; Ho et al., 2015). In contrast, Treg have higher NAD to compensate, and are not affected nearly as much by reduced glycolytic activity (Beier et al., 2015; Gerriets et al., 2015).
We have shown that Foxp3 can mediate the transcriptional repression of Myc, suppress glycolysis, and cause Tregs to acquire resistance to the suppressive effects of L-lactate. We suggest that these metabolic adaptations in Treg could be a mechanism of peripheral tolerance, in which vulnerable ischemic tissues are somewhat protected by a milieu of low glucose and elevated lactate levels, which favors Treg. Treg sustain L-lactate exposure very well, without loss of function, and even increased iTreg cell formation. In contrast, effector and cytotoxic T cells are profoundly impaired by L-lactate (Calcinotto et al., 2012; Fischer et al., 2007). It is possible to speculate that defects in Treg metabolism, and ultimately loss of the Treg metabolic advantage in ischemic tissues can lead to autoimmune diseases such as systemic lupus erythematosus (MacIver et al., 2013).
In addition to preventing autoimmunity, the role of L-lactate metabolism in controlling immunity may be important for tolerance to commensal bacteria such as lactobacilli and bifidobacteria, which produce lactic acid and are known for limiting gastrointestinal inflammation (Poutahidis et al., 2013).
Unfortunately, the Foxp3-mediated adaptations of Treg may also represent an obstacle to anti-cancer immunity. Solid tumors can deprive the tumor microenvironment of glucose and enrich it with lactic acid, which selectively weakens cytotoxic and effector T cells against cancer (Chang et al., 2013; Chang et al., 2015; Ho et al., 2015; Macintyre et al., 2014; Singer et al., 2011), but, based upon the data shown in this work, may leave Treg relatively unscathed, and perhaps contribute to the relative dominance of suppressive Treg cells in the tumor microenvironment. In this way, cancers can hijack a physiologic mechanism of self-tolerance. The tumor microenvironment surrounding many solid cancers has been suggested to be a reason why solid tumors are much less susceptible to chimeric antigen receptor T cell therapy than hematologic malignancies (Newick et al., 2016). The concept that Treg are metabolically better adapted to the tumor microenvironment adds an additional problem to the list of pathologies caused by cancer metabolism, but it also offers new approaches to therapy. It may be possible to protect effector T cells from the suppressive effects of L-lactate by inhibiting LDH. LDH is already a promising drug target to interfere with cancer metabolism (Doherty and Cleveland, 2013; Xie et al., 2014). Another approach maybe to enhance OXPHOS metabolism in engineered chimeric antigen receptor T cells, which elevates their metabolic robustness of the attacking immune cells in the tumor microenvironment (Kawalekar et al., 2016). Either approach may negate the detrimental metabolic advantage of Treg over effector and cytotoxic T cells, and thereby enhance anti-tumor immunotherapy.
Lastly, the model of glucose depriving, lactate producing cancers creating an environment in which Treg are favored could also inspire a novel approach to therapeutic immunosuppression. Organ transplantation offers a unique opportunity to treat the allograft prior to implantation, e.g. to induce a metabolic phenotype similar to cancer metabolism prior to implantation into the recipient, and create a transplant microenvironment mimicking aspects of the tumor microenvironment. It may be possible to reverse engineer the mechanisms of cancer immune escape, and utilizing them for a novel approach to transplant immunosuppression.
In conclusion, we have shown that the Treg transcription factor Foxp3 regulates T cell metabolism, suppresses Myc signaling and glycolysis, enhancing OXPHOS and NAD regeneration. Treg are resistant to the suppressive effects of L-lactate on effector T cells, which are LDH dependent and lead to NAD depletion. These findings represent metabolic adaptations allowing Treg to function in low glucose, high lactate environments, and may be important to peripheral immune tolerance in ischemic tissues, but also weaken anti-cancer immunity in the tumor microenvironment. Our findings may lead to the novel approaches to selectively promote immune modulation in cancers and autoimmune diseases.
STAR★Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed and will be fulfilled by the Lead Contact, Ulf H. Beier (beieru@email.chop.edu).
Experimental Model and Subject Details
Mice
We purchased B6/Rag1−/ −, C57BL/6 (Thy1.1 and Thy1.2) and BALB/c from The Jackson Laboratory (Bar Harbor, ME). We housed Foxp3creYFP (Rubtsov et al., 2008), HDAC6KO (Zhang et al., 2008), as well as mtND6mut (Lin et al., 2012) and mtCO1mut (D.C.W.) from their respective developers, and bred appropriate age and gender matched wild type controls. Mice housed under specific-pathogen-free conditions were studied using protocols approved by the Institutional Animal Care and Use Committees of the Children’s Hospital of Philadelphia and University of Pennsylvania (13–000561 and 14-000746). All animals reproduced at expected Mendelian ratios and were housed under standard conditions (group housing up to five per cage), except B6/Rag1−/− which require high barrier housing (also group housing up to five per cage). Detailed immune phenotyping of HDAC6KO mice has been previously published (Beier et al., 2012; de Zoeten et al., 2011), and is shown in Figures 3 and S4 for mtND6mut and mtCO1mut mice. In vivo mouse models include colitis, cardiac allografting, and homeostatic proliferation. Experimental design, age and gender, and group assignments are explained in the corresponding Method Details sections below.
Cell lines
293T (CRL-3216™) and Phoenix-ECO cells (CRL-3214™) were obtained from American Type Culture Collection (ATCC), both suggested to be female in origin.
Cell culture media and cell culture conditions
For standard cell culture medium, we used RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 U × ml−1), streptomycin (100 mg × ml−1), and 55 nM β2-mercaptoethanol. For low-glucose media, we used substituted RPMI 1640 with glucose-free RMPI-1640 (#11879020) and FBS with glucose free dialyzed FBS (#26400036) from Thermo Fisher Scientific (Waltham, MA). Cells were cultured at 37° C with 5% CO2, except for cells plated prior to seahorse assays, which requires incubation without CO2.
Method Details
Bioenergetic measurements
We measured T cell bioenergetic functions, OCR and ECAR using a XF24 and XF96 analyzer (Seahorse Biosciences, North Billerica, MA), and followed published protocols (Beier et al., 2015; van der Windt et al., 2016). XF24 or XF96 plates were coated using CellTak (BD Biosciences). Isolated T cells were plated in unbuffered XF Assay Media, and then incubated for 10 min at 37° C without CO2. We used 1 × 106 cells per well for XF24, and 2 × 105 cells per well for XF96 assays. To enhance cell adherence, plates were spun at room temperature for 5 min at 400 g. Three baseline measurements of OCR and ECAR were taken and, then, the cells were exposed to assess either a OCR (with final concentrations of 1.25 μM oligomycin, 0.5 μM FCCP, 1 μM rotenone and 1.8 μM antimycin A), or ECAR (with final concentrations of 10 mM glucose, 2 μM oligomycin, and 50 mM 2-Deoxy-D-glucose). Three readings were taken after each sequential injection. Instrumental background was measured in separate control wells using the same conditions without biological material. Seahorse reagents from Agilent Technologies (Wilmington, DE).
Cardiac Allografting
For cardiac allografting, we transplanted BALB/c hearts (H-2d) into the abdomen of C57BL/6 recipients (H-2b). We used female recipients and donors between 8–12 weeks of age, randomly assigned to treatment groups. We chose female mice to minimize fight injuries and age 8–12 to enable sufficiently large blood vessels to enable microsurgeries. Recipients received low-dose rapamycin (0.2 mg × kg−1 × day−1) and either 9 μmol × g−1 Na L-lactate or NaCl, administered intraperitoneally at 150 mM (i.p.) for 14 days from engraftment. Allograft survival was assessed by daily palpation, and rejection was confirmed by histology.
Colitis
We assessed colitis induced using 5% dextran sulfate sodium (DSS, wt/vol, with a molecular weight 36–50 kDa, Cat. #160110, MP Biomedicals, Solon, OH), using a previously published chronic colitis model involving multiple repetitive cycles of 5% DSS designed to asses Th1 based immunopathology (Akimova et al., 2014; Okayasu et al., 1990). Briefly, 12-week-old female C57BL/6 mice (5 per group, randomly selected, and confirmed to have near equal starting weight with no significant difference between groups) received cycling doses of 5% DSS interchanged with normal drinking water every five to seven days (see Figure S6C for details), for a total observation period of 40 days. Female mice were chosen to eliminate fight wounds between animals potentially compromising inflammatory readouts, and a uniform age of 12 weeks is required to exclude somatic weight gain and standardize weight loss observations. Weight and colitis scoring was assessed every weekday. Mice were offered drinking water containing either 150 mM (–) Methyl L-lactate or water control as treatment throughout the experiment.
Cell isolation and flow cytometry
Spleen and peripheral lymph nodes were harvested and processed to single cell suspensions of lymphocytes. We used magnetic beads (Miltenyi Biotec, San Diego, CA) for isolation of Tconv (CD4+CD25−), Treg (CD4+CD25+), and antigen presenting cells (CD90.2−). For cell sorting, we isolated lymphocytes from Foxp3creYFP mice and purified CD4+ cells as above. Then, we sorted CD4+YFP+(Foxp3+) and CD4+YFP− cells via a FACS Aria cell sorter (BD Bioscience, UPenn Cell Sorting Facility). Cells of interest were analyzed using surface markers, and for Foxp3 staining, surface marker-stained cells were fixed, permeabilized, and labeled with Foxp3-specific mAb (Tao et al., 2005). All flow cytometry data was captured using Cyan (Dako) as well as Cytoflex (Beckman Coulter, Brea, CA) and analyzed using the FlowJo 10.1r5 software. Pooled histogram data are shown as percent of maximum (% of max), which is a normalization of overlaid data and represents number of cells in each bin divided by the number of cells in the bin that contains the largest number of cells.
Chromatin Immunoprecipitation
Using Magna ChIP A/G Kits (#17-408, Merck Millipore, Billerica, MA) and Pierce Magnetic ChIP kits (ThermoFisher Scientific, Waltham, MA), DNA-protein complexes were immunoprecipitated from 2 million Tregs per condition using either isotype control IgG or anti-Foxp3 antibody (eBioscience cat# 14-4774-82). The immunoprecipitated genomic DNA was probed by RT-PCR using primers for Myc promoter: forward 5′-TGA TCA GGG CCG ACT TTT TT-3′ and reverse 5′-AAC CCA AGC TTT CCC CTT TTA TT-3′; Myc TATA box region: forward 5′-TAG CGC GCG AGC AAG AG-3′ and reverse: 5′-GTA AAC AGT AAT AGC GCA TGA AT-3′.
ChIP-on-chip
For our ChIP-chip assays, we used NimbleGen Mouse ChIP-chip 3 × 720K RefSeq Promoter Arrays (NimbleGen Roche, Madison, WI). Briefly, we isolated three independent samples of gender and age matched HDAC6KO and WT control CD4+CD25+ Treg, isolated, cross-linked and processed genomic DNA as above. DNA-protein complexes were bound with Foxp3 mAb, and enriched through immunoprecipitation, which a control sample is retained without immunoprecipitation (input). Both input and Foxp3-IP enriched samples underwent whole-genome amplification or ligation mediated-PCR to obtain adequate amounts of DNA for labeling. Foxp3-IP enriched samples were labeled with Cy5, and input samples with Cy3, and the arrays submitted to NimbleGen for analysis. Data were analyzed using Partek GS (Partek Inc., St Louis, MO) and the Broad Institutes Integrative Genomics Viewer (http://software.broadinstitute.org/software/igv/, Version 2.3.32) (Robinson et al., 2011). Raw data underwent Loess normalization, normal distribution testing, and Student t-testing with a false discovery rate of 0.1 (p-value cutoff 0.0029). For visualization in the Broad Institutes Integrative Genomics Viewer, we generated IGV files using NimbleGen mapping information from the SignalMap GFF files.
Electron microscopy
We used processed and analyzed electron microscopy samples as previously described (Beier et al., 2015). Briefly, purified HDAC6KO and C57BL/6 Tconv and Treg were centrifuged and fixed in 2.5% glutaraldehyde in 0.1 M Sorenson’s phosphate buffer. Following fixation pellets were treated as follows: 3 × 10 min changes of 0.1 M Sorenson’s phosphate buffer adjusted to 425 mOsm × L−1 with sucrose followed by fixation in 1% osmium tetroxide in 0.1 M Sorenson’s phosphate buffer for 1.5 h; washed in de-ionized water (3 × 10 min) followed by en bloc staining with 2% uranyl acetate for 30 min; dehydration in acetone; infiltration and embedding with increasing concentrations of Spurr’s resin in acetone. Ultrastructural images were visualized using a Philips EM208S transmission electron microscope by a pathologist blinded to experimental conditions (T.R.B.). The number of mitochondria present in each cell (24/sample, 11,000 to 22,000-fold magnification) were recorded. Cells without intact nuclei were excluded from analysis to minimize inclusion of changes secondary to preservation artifact or cellular degeneration.
Glucose uptake by flow cytometry
For measuring glucose uptake in T cells, we used 2-NBDG (2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose, purchased from Cayman Chemicals, Ann Arbor, MI), based upon a previously published protocol (Sukumar et al., 2013). Briefly, we diluted 2-NBDG into glucose free media to 150 μg × ml−1. After cell culture and stimulation in a 96-well plate, the stimulation media was removed, and the 2-NBDG containing glucose free media added (200 μl), avoiding light exposure. Subsequently, the cells incubated for 10 min (37° C, 5% CO2), and then the 2-NBDG staining solution removed, before the cells were processed by flow cytometry (without fixation and permeabilization, and minimizing light exposure). Unstained cells undergoing the same stimulation served as negative controls.
Histology
Cardiac allografts were fixed in 10% neutral buffered formalin, routinely processed and embedded in paraffin. Histologic sections for light microscopy were cut to a thickness of 4 μm and stained with hematoxylin and eosin (H&E) and were reviewed by a pathologist (T.R.B.) blinded to treatment conditions. For immunoperoxidase staining, tissues were fixed with 2% paraformaldehyde, rinsed in buffer, pretreated with 10% normal goat serum and 0.1% Triton X-100 (Sigma-Aldrich), and incubated overnight with rat mAbs to CD3 (cat. # A0452, Dako) and Foxp3 (FJK-16s, eBioscience) diluted in 1% normal goat serum and 0.1% Triton X-100. Sections were washed and incubated with rabbit anti-rat IgG (Dako) plus 0.3% H2O2 in methanol (4°C, 10 min) to block endogenous peroxidase, and bound antibodies were detected using peroxidase-conjugated anti-rabbit IgG and diaminobenzidine substrate (Envision kit; Dako). Immunostained slides were scanned using the Aperio ScanScope® CS slide scanner (Aperio Technologies, Vista, CA). Whole slide digitized images were analyzed using the Aperio ImageScope software (version 10.0.1346.1807; Aperio Technologies, Vista, CA) for determination of the percentage of cells with membranous decoration for CD3 and Foxp3 from the total number of cells present on the slide.
Homeostatic proliferation
To assess the effect of Na L-lactate on T cell function in vivo, we adoptively transferred 1 × 106 CD90.1+ CD4+ T effector cells without or without 2.5 × 105 CD90.2+CD4+ Tregs into 8-week-old female B6/RAG1−/− mice. Mice were randomly assigned to the treatment groups, either 9 μmol × g−1 Na L-lactate or NaCl, administered intraperitoneally at 150 mM (i.p.) for seven days (Figure 5G), or, 9 μmol × g−1 Methyl L-lactate administered i.p. at 150 mM, using equal volume H2O injection as control (Figure S7B). Female mice were chosen to prevent fight wounds in the B6/RAG1−/− mice potentially compromising inflammatory readouts, while the age of 8 weeks. Eight weeks was chosen as it balances the feasibility of tail vein injections (more difficult with younger mice) with cost (higher with older mice). Splenocytes were obtained one week later, quantified, and CD90.1, CD4 and Foxp3 were assessed by flow cytometry as previously reported (Beier et al., 2012).
Immunoblotting
Immunoblotting was performed as previous reported (Beier et al., 2012). Purified cells of interested were lyzed in radioimmunoprecipitation assay (RIPA) buffer with Halt protease inhibitor (Thermo Fisher Scientific). Protein concentration was determined by photometry (DU640, Beckman-Coulter). Subsequently, samples were mixed with Laemmli sample buffer containing 2-mercaptoethanol (Bio-Rad Laboratories, Hercules, CA) and boiled for 5 min, except samples subjected to Mitoprofile immunoblotting, which should not be boiled during processing (Beier et al., 2015). The protein samples were loaded onto Mini-PROTEAN TGXTM 4 to 15% gradient gels (Bio-Rad) and underwent electrophoretic separation. Proteins were then transferred to PolyScreen PVDF Hybridization Transfer Membranes (PerkinElmer, Waltham, MA). Membranes were cut according to the molecular weights of the proteins of interest (as indicated with Precision Plus Protein Dual Color, Bio-Rad) and incubated with primary and horseradish peroxidase (HRP)–conjugated secondary antibodies. We used Super Signal West Pico chemiluminescent substrate (Thermo Fisher Scientific) and X-OMAT Blue XB Film (Kodak, Rochester, NY) for image development. Depending on the background, films were scanned or photographed and then processed with Adobe Photoshop CC 2016 (grayscale conversion and auto contrast function). Densitometric analysis was performed using ImageJ64 version 1.45S (https://imagej.nih.gov/ij/).
Measurement of glucose, L-lactate and D-lactate
Glucose was measured by using a Contour® next EZ glucometer system (Bayer, Robinson Township, PA). We measured L- and D-lactate through a fluorescence-based assay with recombinant LDH and LDHD (Cayman Chemical, Ann Arbor, MI; cat. #700510 and #700520, respectively) following the manufacturers’ instructions. In the reaction, L-lactate or D-lactate is determined by the oxidation of lactate to pyruvate, and the reduction of NAD to NADH, with NADH binding to a fluorescent substrate. The samples were incubated for 20 min (L-lactate) or 30 min (D-lactate) at room temperature and then measured with a Spectramax Gemini XPS plate reader using an excitation wavelength of 530–540 nm and reading an emission wave length of 585–595 nm. Data were analyzed using SoftMax Pro 4.7 software (Molecular Devices, Sunnydale, CA), MS Excel, and Prism. We used these assays to assess LDH isoform specificity of the LDH inhibitor GSK 2837808A (Tocris Bioscience, Bristol, UK) (Billiard et al., 2013; Xie et al., 2014). Briefly, lactate standard curve samples were prepared in duplicate (final concentrations: 0, 25, 50, 100, 200, 400, 600, and 1,000 μM). We prepared investigational samples at 600 μM sodium L-lactate or D-lactate (Sigma-Aldrich) dissolved in assay buffer, with or without vehicle control (DMSO) and incremental doses of the LDH inhibitor.
Metabolite extraction and derivatization
T cells were counted and volumes determined using a Multisizer 3 Counter Counter (Beckman Coulter, Pasadena, CA). Cells were washed twice with phosphate-buffered saline (PBS) followed by scraping into 750 μl of ice-cold methanol/water (4:1, v/v) containing 1 mg of Phenylhydrazine hydrochloride for derivatization. Samples were pulse-sonicated for 30 seconds and centrifuged at 16,000 g for 10 min. The supernatant was then transferred to a clean tube and samples were incubated at for 1 hour at room temperature. Following incubation, samples were evaporated to dryness under nitrogen and suspended in 60 μL of water prior to liquid chromatography mass spectrometry analysis.
Microarrays
Microarray experiments were performed using whole-mouse-genome oligoarrays (Mouse430a; Affymetrix), and array data were analyzed using MAYDAY software (http://it.inf.uni-tuebingen.de/?page_id=248) version 2.12 (Battke et al., 2010). Array data were subjected to robust multiarray average normalization. To assess differential gene expression, fold changes of up- and downregulated genes were calculated, and significance assessed via Significance Analysis of Microarrays (SAM) using 100 permutations and false discovery rate <0.1, and data with >1.2-fold differential expression were included in the analysis. Data underwent z-score transformation for heatmap display.
NAD and NADH measurement
Nicotinamide adenine dinucleotide (NAD) and Nicotinamide adenine dinucleotide hydrate (NADH) were measured in heart and liver isolated from wild type (WT), ND6mut and COImut mice (4–6 per group) using the NAD:NADH Quantification Kit (Sigma-Aldrich MAK037). The tissues were snap-frozen in liquid nitrogen to prevent the NAD and NADH consumption by enzymes. Then, the tissues (20–25 mg) were homogenized in NADH:NAD extraction buffer (350–400 μl) in ice and spun at 14,000 g for 5 min at 4° C, afterward the supernatant was transferred in 10 kDa molecular weight cut off spin filter and spun at 14,000 g for 5 min at 4° C to be deproteinized. The supernatant was divided in two aliquots, one aliquot was kept in ice and used for NADtotal measurement (defined as the sum of oxidized NAD+ and reduced NADH), and the other aliquot was incubated at 60° C for 30 min to decompose NAD leaving NADH only. The samples were transferred to a 96 wells plate and the absorbance at 450 nm (A450) was measured using a SpectraMax Paradigm Multi-Mode Detection Platform (Molecular Devices). The background was subtracted from the measurement and the amount of NADtotal and NADH present in the samples was determined from the standard curve. The amount of oxidized NAD (NAD+) was obtained subtracting NADH from NADtotal. For measurement of NAD and NADH in iTreg, we isolated CD4+CD25− Tconv from Foxp3YFPcre mice, and co-stimulated and polarized the cells to form iTreg (see supplemental figure S1A, Fig 1A). After 4 days, the iTregs were separated from the cells that did not express Foxp3 (‘non-iTregs’) through FACS. Importantly, our preliminary studies had shown that washing and sorting T cells nullifies the effects of in culture conditions on NAD:NADH polarization. Therefore, after cell sorting, we re-exposed the iTreg and non-iTreg samples to culture at 37° C and 5% CO2 and T cell culture medium with platebound CD3ε/CD28 mAb and 25 U × mL−1 IL-2 for an hour prior to harvesting the cells for NAD/NADH measurement.
T cell isolation and function
For Treg suppression assays, purified Tconv cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) or CellTrace Violet (both ThermoFisher) and stimulated with irradiated antigen presenting cells plus CD3ε mAb (1 μg × ml−1, BD Pharmingen). After 72 h, proliferation of Tconv cells was determined by flow cytometric analysis of CFSE or CellTrace dilution, followed by normalization of relative suppression and calculation of area-under-curve (Akimova et al., 2016). For conversion to Foxp3+ Tregs, Tconv cells were incubated for 3–5 days with CD3ε/CD28 mAb beads, plus TGF-β (3 ng × ml−1) and IL-2 (25 U × ml−1), and analyzed by flow cytometry for Foxp3+ iTreg (de Zoeten et al., 2010). For in vivo Treg expansion, IL-2/anti-IL-2 complexes were prepared by incubating 2 μg recombinant mouse IL-2 (Biolegend, San Diego, CA) with 10 μg anti-IL-2 mAb (Bioxcell, West Lebanon, NH) in 200 μl PBS for 30 min at 37° C. Mice were injected, intraperitoneally, for three days and spleen and LNs were harvested on day 5 for Treg isolation.
T cell transduction
Foxp3 retroviral transductions were conducted as previously described (Chen et al., 2006; Liu et al., 2012). Retroviruses were generated by cotransfection of MinR1-Foxp3 vector or parental MinR1 vector (empty vector, EV), plus pCLeco (Invitrogen, CA) helper plasmid into the 293T-based Phoenix ecotropic packaging cell line (Phoenix-ECO, CRL-3214™), using Lipfectamine 2000 (11668-019, Invitrogen). Virus containing supernatant was used to inoculate magnetically purified C57BL/6 CD4+ T cells (130-049-201, Miltenyi Biotec). Isolated CD4+ T cells were stimulated for 20 h using phorbol 12-myristate 13-acetate (PMA, 3 ng/mL), ionomycin (1 μM) and mouse IL-2 (10 U × mL−1, Roche). Activated T cells were infected with 48 h and 72 h viral supernatants harvested from transfected Phoenix cells, plus 10 U × mL−1 mouse IL-2 and 4 μg × mL−1 Polybrene (Sigma-Aldrich), and centrifuged for 90 min at 3,200 rpm (1533.67 g, Eppendorf 5810R). Cells cultured at 37° C with 5% CO2 for 3 days were used in seahorse bioenergetic assays. Transduction efficiency was confirmed by flow cytometry and documentation of gain of suppressive function in Foxp3, but not EV transduced CD4+ T cells.
Th17 polarization
We assessed Th17 polarization as previously described (Thomas et al., 2012; Xiao et al., 2016). Briefly, C57BL/6 splenocytes were depleted of CD8+ cells using anti-CD8 microbeads, and the remaining cells were cultured with soluble CD3ε and CD28 mAb (1 μg × ml−1 each) for four days in the presence of anti-IL-4 and anti-IFN-γ mAbs (20 μg × ml−1), TGFβ (1 ng × ml−1), and IL-6 (10 ng × ml−1). For intracellular IL-17 staining, cells were stimulated with 30 ng × ml−1 phorbol 12-myristate 13-acetate and 1 μM ionomycin (Sigma-Aldrich) for 5 h in the presence of Golgi Stop (BD Biosciences).
Total ATP measurement
The total amount of ATP has been measured in spleen, heart, liver isolated from WT, ND6 and COI mice (4–6 per group) using the ATP Fluorimetric Assay Kit (Sigma-Aldrich MAK190). The tissues were snap-frozen in liquid nitrogen to prevent the ATP consumption by enzymes. Then, the tissues (10–20 mg) were homogenized in ATP assay buffer (300–400 μl) in ice and spun at 14,000 g for 5 min at 4° C, afterward the supernatant was transferred in 10 kDa molecular weight cut off spin filter and spun at 14,000 g for 5 min at 4° C to be deproteinized. The samples were transferred to a 96 wells plate and the fluorescence (λex= 535 nm, λem= 587 nm) was measured using a SpectraMax Paradigm Multi-Mode Detection Platform (Molecular Devices). The background was subtracted from the measurement and the amount of ATP present in the samples was determined from the standard curve.
RNA analysis
RNA was extracted using RNeasy kits (Qiagen, Hilden, Germany), and RNA integrity and quantity were analyzed by photometry (Nanodrop 2000, Thermo Fischer). Revere transcription and qPCR were performed as reported (Beier et al., 2015; Beier et al., 2011; Beier et al., 2012). Isolated RNA was reverse transcribed to cDNA with random hexamers and amplified (PTC-200; MJ Research). Primer sequences for target genes were used for quantitative PCR amplification of total cDNA. All primers were purchased from Applied Biosystems. Differences in cDNA input were corrected by normalizing signals obtained with specific primers for 18S rRNA. Relative quantitation of target cDNA was determined by the formula 2−ΔCT, with ΔCT denoting fold increases above the set control value (resting Tconv). Data was analyzed using StepOnePlusTM (Applied Biosystems), Excel, and Prism.
Ultra-high-performance liquid chromatography-selected reaction monitoring/mass spectrometry
We co-stimulated Treg and Tconv with CD3ε/CD28 mAb-coated beads and 25 U IL-2 × ml−1 for 2 hours in T cell medium with either 200 mg × dl−1 D-glucose and added 20 mM NaCl (control media), or with 20 mg × dl−1 D-glucose with 20 mM Na L-lactate (low glucose high lactate media, LGHL). For labeling, we replaced [13C6]-D-glucose in the control media, and Na [13C3]-L-lactate in the LGHL media, respectively. We purchased [13C6]-D-glucose (CLM-1396-PK) and sodium [13C3]-L-lactate (CLM-1579-PK) from Cambridge Isotope Laboratories (Tewksbury, MA). We continued co-stimulation for an additional 16 hours under the same conditions. Metabolite isolation and ultra-high-performance liquid chromatography-selected reaction monitoring/mass spectrometry is based upon previously published methodology (Guo et al., 2016), as following. Organic acids from cell samples were analyzed using an Agilent 1200 series high performance liquid chromatography system coupled to an Agilent 6460 triple quadrupole mass spectrometer equipped with an electrospray ionization source operated in negative ion mode. Analytes were separated by reversed-phase ion-pairing chromatography utilizing a Waters Atlantis T3 column (150 μm × 2.10 mm, 3 μm particle size) at a flow rate of 200 μL × min−1 maintained at 50° C. A two-solvent gradient system was used with solvent A as 200 mM 1,1,1,3,3,3-hexafluoro-2-propanol and 5 mM N-Ethyl-N-(propan-2-yl)propan-2-amine (DIPEA) in water and solvent B as 200 mM 1,1,1,3,3,3-hexafluoro-2-propanol and 5 mM DIPEA in methanol. The linear gradient conditions were as follows: 1% B at 0 min, 1% B at 6 min, 40% B at 13 min, 95% B at 18 min, 95% B at 24.5 min, 1% B at 27 min followed by a 5-min equilibration. The Agilent 6460 mass spectrometer operating conditions were as follows. The gas temperature was set at 325° C, and the gas flow was set to 8 liters × min−1. The sheath gas temperature was 400° C, and the sheath gas flow was set to 10 liters per min. The capillary voltage was set to 3,000 V, and the L nozzle voltage was set to 1,000 V. The isotopic distribution of tricarboxylic acid cycle metabolites was calculated as described previously (Fernandez et al., 1996).
Quantification and Statistical Analysis
Statistical analysis was conducted using GraphPad Prism 6 software. All data were tested for normal distribution of variables. All normally distributed data were displayed as means ± standard error of the mean (SEM) unless otherwise noted. Measurements between two groups were performed with an unpaired Student-t test if normally distributed, or Mann-Whitney U test if otherwise. For paired samples, we used a paired Student-t test or Wilcoxon matched-pairs signed rank test, depending on whether data were normally distributed, respectively. Groups of three or more were analyzed by one-way analysis of variance (ANOVA) or the Kruskal-Wallis test. Allograft survival was assessed using a log-rank (Mantel-Cox) test. Statistical parameters for each experiment can be found within the corresponding figure legends.
Supplementary Material
Figure S1: Oxygen consumption in naïve and stimulated Tconv and Treg. Related to Figure 1.
Figure S2: HDAC6 deletion and inhibition increases T cell oxygen consumption. Related to Figure 1 and 2.
Figure S3: Electron transport chain complexs in Tconv and Treg. Related to Figure 2.
Figure S4: Loss of mitochondrial ND6 and CO1 leads to reduced NAD oxidation and ATP production, respectively, without defects in T cell development. Related to Figure 3.
Figure S5: Tricaboxylic acid derivatives in activated effector and regulatory T cells. Related to Figure 4.
Figure S6: L-lactate does not induce apoptosis or NRF-2 signaling. Related to Figure 5 and 6.
Figure S7: Methyl L-lactate does not suppress T cell function in vivo, and GSK 2837808A does not inhibit LDHD. Related to Figure 6 and 7.
Acknowledgments
We thank Roddy S. O’Conner and Michael C. Milone (University of Pennsylvania) for advice and help with measuring T cell volumes and Haiyan Xiao (Children’s Hospital of Philadelphia) for technical assistance. We thank Eric Rappaport (Children’s Hospital of Philadelphia) and Nicolai M. Doliba (University of Pennsylvania) for allowing us to use equipment. We thank Jeffrey C. Rathmell (Vanderbilt University) for helpful comments. Financial support was as follows: AI095353 (to U.H.B.); AI073489 and AI095276 (to W.W.H.); HHMI International Student Research fellowship (to P.K.K.); DK092282 and DK106243 (to M.H.L.); DK101600 (to B.L.L.); OD010944, NS021328, MH108592, and CA182384 (to D.C.W.); and DK098656 (to J.A.B.).
Footnotes
Author Contributions
A.A., L.G.-D.-G., S.D., J.J., U.H.B., Z.W., L.W., T.R.B., P.K.K., T.A., Y.L., R.H., and W.Q. performed experiments. U.H.B., W.W.H., M.H.L., T.A., L.G., L.G.-D.-G., A.A., and S.D. designed experiments. U.H.B., W.W.H., A.A., L.G.-D.-G., S.D., L.G., M.H.L., T.R.B., T.A., W.Q., Y.L., J.A.B., L.W., I.A.B., D.C.W., and B.L.L. analyzed data. U.H.B. wrote the manuscript. B.L.L., T.A., W.Q., J.A.B., W.W.H., A.A., S.D., L.G.-D.-G., and D.C.W. edited the manuscript.
Data and Software Availability
Microarray and genomic data accession numbers
We have deposited our microarray and ChIP-chip data in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). The accession numbers for the data reported in this paper are GEO: GSE27082, GSE92520.
References
- Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PLoS One. 2011;6:e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimova T, Levine MH, Beier UH, Hancock WW. Standardization, Evaluation, and Area-Under-Curve Analysis of Human and Murine Treg Suppressive Function. Methods Mol Biol. 2016;1371:43–78. doi: 10.1007/978-1-4939-3139-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimova T, Xiao H, Liu Y, Bhatti TR, Jiao J, Eruslanov E, Singhal S, Wang L, Han R, Zacharia K, et al. Targeting sirtuin-1 alleviates experimental autoimmune colitis by induction of Foxp3+ T-regulatory cells. Mucosal Immunol. 2014;7:1209–1220. doi: 10.1038/mi.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battke F, Symons S, Nieselt K. Mayday--integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier UH, Angelin A, Akimova T, Wang L, Liu Y, Xiao H, Koike MA, Hancock SA, Bhatti TR, Han R, et al. Essential role of mitochondrial energy metabolism in Foxp3(+) T-regulatory cell function and allograft survival. FASEB J. 2015;29:2315–2326. doi: 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier UH, Wang L, Bhatti TR, Liu Y, Han R, Ge G, Hancock WW. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol Cell Biol. 2011;31:1022–1029. doi: 10.1128/MCB.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier UH, Wang L, Han R, Akimova T, Liu Y, Hancock WW. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci Signal. 2012;5:ra45. doi: 10.1126/scisignal.2002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard J, Dennison JB, Briand J, Annan RS, Chai D, Colon M, Dodson CS, Gilbert SA, Greshock J, Jing J, et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013;1:19. doi: 10.1186/2049-3002-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brea D, Agulla J, Rodriguez-Yanez M, Barral D, Ramos-Cabrer P, Campos F, Almeida A, Davalos A, Castillo J. Regulatory T cells modulate inflammation and reduce infarct volume in experimental brain ischaemia. J Cell Mol Med. 2014;18:1571–1579. doi: 10.1111/jcmm.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcinotto A, Filipazzi P, Grioni M, Iero M, De Milito A, Ricupito A, Cova A, Canese R, Jachetti E, Rossetti M, et al. Modulation of microenvironment acidity reverses anergy in human and murine tumor-infiltrating T lymphocytes. Cancer Res. 2012;72:2746–2756. doi: 10.1158/0008-5472.CAN-11-1272. [DOI] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Rowell EA, Thomas RM, Hancock WW, Wells AD. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J Biol Chem. 2006;281:36828–36834. doi: 10.1074/jbc.M608848200. [DOI] [PubMed] [Google Scholar]
- Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, Bradner JE, Mazitschek R, Kozikowski AP, Matthias P, Hancock WW. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol. 2011;31:2066–2078. doi: 10.1128/MCB.05155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010;138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Hasumi K, Sakai K, Kanbe T. Specific inhibition of glyceraldehyde-3-phosphate dehydrogenase by koningic acid (heptelidic acid) J Antibiot (Tokyo) 1985;38:920–925. doi: 10.7164/antibiotics.38.920. [DOI] [PubMed] [Google Scholar]
- Ewaschuk JB, Naylor JM, Zello GA. D-lactate in human and ruminant metabolism. J Nutr. 2005;135:1619–1625. doi: 10.1093/jn/135.7.1619. [DOI] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- Frattini V, Pisati F, Speranza MC, Poliani PL, Frige G, Cantini G, Kapetis D, Cominelli M, Rossi A, Finocchiaro G, Pellegatta S. FOXP3, a novel glioblastoma oncosuppressor, affects proliferation and migration. Oncotarget. 2012;3:1146–1157. doi: 10.18632/oncotarget.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, Warmoes MO, de Cubas AA, MacIver NJ, Locasale JW, et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016 doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets VA, Kishton RJ, Nichols AG, Macintyre AN, Inoue M, Ilkayeva O, Winter PS, Liu X, Priyadharshini B, Slawinska ME, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Guo L, Shestov AA, Worth AJ, Nath K, Nelson DS, Leeper DB, Glickson JD, Blair IA. Inhibition of Mitochondrial Complex II by the Anticancer Agent Lonidamine. J Biol Chem. 2016;291:42–57. doi: 10.1074/jbc.M115.697516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann P, Boeld TJ, Eder R, Huehn J, Floess S, Wieczorek G, Olek S, Dietmaier W, Andreesen R, Edinger M. Loss of FOXP3 expression in natural human CD4+CD25+ regulatory T cells upon repetitive in vitro stimulation. Eur J Immunol. 2009;39:1088–1097. doi: 10.1002/eji.200838904. [DOI] [PubMed] [Google Scholar]
- Howie D, Cobbold SP, Adams E, Bokum AT, Necula AS, Zhang W, Huang W, Roberts DJ, Thomas B, Hester SS, et al. Foxp3 drives oxidative phosphorylation and protection from lipotoxicity. JCI Insight. 2017;2:e89160. doi: 10.1172/jci.insight.89160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh A, DuPage M, Priyadharshini B, Sage PT, Quiros J, Borges CM, Townamchai N, Gerriets VA, Rathmell JC, Sharpe AH, et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol. 2015;16:188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalekar OU, O’Connor RS, Fraietta JA, Guo L, McGettigan SE, Posey AD, Jr, Patel PR, Guedan S, Scholler J, Keith B, et al. Distinct Signaling of Coreceptors Regulates Specific Metabolism Pathways and Impacts Memory Development in CAR T Cells. Immunity. 2016;44:380–390. doi: 10.1016/j.immuni.2016.01.021. [DOI] [PubMed] [Google Scholar]
- Lee CF, Lo YC, Cheng CH, Furtmuller GJ, Oh B, Andrade-Oliveira V, Thomas AG, Bowman CE, Slusher BS, Wolfgang MJ, et al. Preventing Allograft Rejection by Targeting Immune Metabolism. Cell Rep. 2015;13:760–770. doi: 10.1016/j.celrep.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CS, Sharpley MS, Fan W, Waymire KG, Sadun AA, Carelli V, Ross-Cisneros FN, Baciu P, Sung E, McManus MJ, et al. Mouse mtDNA mutant model of Leber hereditary optic neuropathy. Proc Natl Acad Sci U S A. 2012;109:20065–20070. doi: 10.1073/pnas.1217113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang L, Han R, Beier UH, Hancock WW. Two lysines in the forkhead domain of foxp3 are key to T regulatory cell function. PLoS One. 2012;7:e29035. doi: 10.1371/journal.pone.0029035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, Rathmell JC. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok CC, Lau CS. Pathogenesis of systemic lupus erythematosus. J Clin Pathol. 2003;56:481–490. doi: 10.1136/jcp.56.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DL. Mechanisms maintaining peripheral tolerance. Nat Immunol. 2010;11:21–27. doi: 10.1038/ni.1817. [DOI] [PubMed] [Google Scholar]
- Newick K, Moon E, Albelda SM. Chimeric antigen receptor T-cell therapy for solid tumors. Mol Ther Oncolytics. 2016;3:16006. doi: 10.1038/mto.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. Int J Cancer. 2010;127:759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Curr Opin Immunol. 2014;27:1–7. doi: 10.1016/j.coi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- Poutahidis T, Kleinewietfeld M, Smillie C, Levkovich T, Perrotta A, Bhela S, Varian BJ, Ibrahim YM, Lakritz JR, Kearney SM, et al. Microbial reprogramming inhibits Western diet-associated obesity. PLoS One. 2013;8:e68596. doi: 10.1371/journal.pone.0068596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Singer K, Gottfried E, Kreutz M, Mackensen A. Suppression of T-cell responses by tumor metabolites. Cancer Immunol Immunother. 2011;60:425–431. doi: 10.1007/s00262-010-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- Tao R, Wang L, Han R, Wang T, Ye Q, Honjo T, Murphy TL, Murphy KM, Hancock WW. Differential effects of B and T lymphocyte attenuator and programmed death-1 on acceptance of partially versus fully MHC-mismatched cardiac allografts. J Immunol. 2005;175:5774–5782. doi: 10.4049/jimmunol.175.9.5774. [DOI] [PubMed] [Google Scholar]
- Thomas RM, Sai H, Wells AD. Conserved intergenic elements and DNA methylation cooperate to regulate transcription at the il17 locus. J Biol Chem. 2012;287:25049–25059. doi: 10.1074/jbc.M112.351916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, Chang CH, Pearce EL. Measuring Bioenergetics in T Cells Using a Seahorse Extracellular Flux Analyzer. Curr Protoc Immunol. 2016;113:3 16B 11–13 16B 14. doi: 10.1002/0471142735.im0316bs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J, Brunen D, Fleskens V, Pals CE, Lam EW, Coffer PJ. Rapid temporal control of Foxp3 protein degradation by sirtuin-1. PLoS One. 2011;6:e19047. doi: 10.1371/journal.pone.0019047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JS, Strayer DS. Immunopathology. In: Rubin EL, Reisner H, editors. Essentials of Rubin’s Pathology. 2013. pp. 85–86. [Google Scholar]
- Xiao H, Jiao J, Wang L, O’Brien S, Newick K, Wang LC, Falkensammer E, Liu Y, Han R, Kapoor V, et al. HDAC5 controls the functions of Foxp3(+) T-regulatory and CD8(+) T cells. Int J Cancer. 2016;138:2477–2486. doi: 10.1002/ijc.29979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, Signoretti S, Billiard J, Duffy KJ, Grant A, et al. Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19:795–809. doi: 10.1016/j.cmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Oxygen consumption in naïve and stimulated Tconv and Treg. Related to Figure 1.
Figure S2: HDAC6 deletion and inhibition increases T cell oxygen consumption. Related to Figure 1 and 2.
Figure S3: Electron transport chain complexs in Tconv and Treg. Related to Figure 2.
Figure S4: Loss of mitochondrial ND6 and CO1 leads to reduced NAD oxidation and ATP production, respectively, without defects in T cell development. Related to Figure 3.
Figure S5: Tricaboxylic acid derivatives in activated effector and regulatory T cells. Related to Figure 4.
Figure S6: L-lactate does not induce apoptosis or NRF-2 signaling. Related to Figure 5 and 6.
Figure S7: Methyl L-lactate does not suppress T cell function in vivo, and GSK 2837808A does not inhibit LDHD. Related to Figure 6 and 7.