Abstract
In vivo interrogation of the function of genes implicated in tumorigenesis is limited by the need to generate and cross germline mutant mice. Here we describe approaches to model colorectal cancer (CRC) and metastasis that rely on in situ gene editing and orthotopic organoid transplantation in mice without cancer predisposing mutations. Autochthonous tumor formation is induced by CRISPR–Cas9-based editing of the Apc and Trp53 tumor suppressor genes in colon epithelial cells and by orthotopic transplantation of Apc-edited colon organoids. ApcΔ/ Δ;KrasG12D/+;Trp53Δ/ Δ (AKP) mouse colon organoids and human CRC organoids engraft in the distal colon and metastasize to the liver. Finally, we apply the orthotopic transplantation model to characterize the clonal dynamics of Lgr5+ stem cells and demonstrate sequential activation of an oncogene in established colon adenomas. These experimental systems enable rapid in vivo characterization of cancer-associated genes and reproduce the entire spectrum of tumor progression and metastasis.
Recent tumor sequencing studies have identified a large number of candidate genes that are mutated in CRCs and may contribute to carcinogenesis, tumor phenotype, and treatment responses in subsets of patients.1,2 Traditionally, functional assessment of putative cancer-associated genes in vivo has required the development of genetically engineered mouse models (GEMMs) of CRC through extensive intercrossing or de novo generation of gene-targeted mice, which is expensive and time consuming. Most GEMMs of CRC such as the ApcMin mouse3,4 are also limited by delayed tumor onset (i.e., 2–4 months) and high tumor burden (i.e., 30–100 polyps) in the small intestine, which is a rare location for human intestinal tumors and precludes study of tumor progression beyond early adenomas or longitudinal studies using colonoscopy5,6. Tumorigenesis can be localized to the colon with either Apc loss driven by a colon-specific promoter, which is limited by slow tumor growth (i.e., 4–6 months)7–9, or somatic deletion of Apc in the distal colon of Apcfl/fl mice with rectal enema of adenoviral Cre, which is requires colonic injury and/or time-consuming surgery10–12.
In addition to GEMMs, human and mouse cell lines are used to model CRC in vivo. Typical sites of transplantation are the mouse flank or kidney capsule, which do not recapitulate the native stroma of the colon mucosa.6 Several groups have sought to orthotopically deliver tumor cell lines into the mouse colon, either surgically into the cecal serosa13 (which is not the relevant tissue layer for CRC development) or into the mucosa via rectal enema14, injury15, electrocoagulation16, or colonoscopy-guided mucosal injection17. However, all of the published orthotopic models are limited by the use of mouse or human cell lines that are not genetically defined and poorly recapitulate the histology of CRC.
The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 nuclease genome editing system offers the ability to somatically mutate one or more genes in wild-type mice to assess their role in tumorigenesis18. We and others have demonstrated the feasibility of inducing tumorigenesis in the lung by lentiviral delivery of CRISPR–Cas9 components19,20. CRISPR–Cas9 gene editing has been applied to engineer human CRC three-dimensional cultures, or organoids; mutations in APC, KRAS, TP53, SMAD4, and/or PIK3CA were required for successful engraftment at ectopic sites in mice21,22.
Here we describe CRISPR–Cas9-based somatic gene editing and orthotopic organoid transplantation approaches that employ colonoscopy-guided mucosal injections for primary and metastatic tumor induction in mice without cancer predisposing mutations. We first optimized a colonoscopy-guided mucosal injection system to produce a mucosal bubble that localizes the injection to the lamina propria of the distal 4 cm of the mouse colon (Supplementary Figure 1a, Supplementary Video 1), based on previous reports17,23. Using this approach, up to three injections per mouse delivering 50–100 μl each can be performed. We observed recombination in a 5-mm diameter circular area at each injection site by bioluminescence and fluorescence in Rosa26LSL-tdTomato/LSL-Luciferase mice receiving an adenoviral vector encoding Cre recombinase (Ad5CMV::Cre), which suggests that our approach specifically infects colon cells at the injection location (Supplementary Figure 1b).
To determine the utility of mucosal injection for viral infection of colonic intestinal stem cells at the crypt base, we injected a lentiviral vector that incorporates the human kinase-1 (PGK) promoter driving the expression of Cre recombinase (lenti-PGK::Cre) into Rosa26LSL-tdTomato/+ mice, and then traced recombined cells by immunofluorescence. We observed infection of epithelial cells in the crypt base (the location of colon stem cells that initiate tumorigenesis). Notably, we also detected widespread recombination in stromal cells of the colon (Supplementary Figure 1c), but did not observe recombination in the proximal colon, small intestine, or liver by fluorescence microscopy (data not shown), indicating that mucosal viral infection of the mucosa is restricted to the epithelial cells and the lamina propria stromal cells of the colon in the injection area.
Consistent with these findings, deletion of the Apc tumor suppressor gene by mucosal injection of the PGK::Cre lentivirus into Apcfl/fl mice reliably produced tumors within six weeks that were monitored by colonoscopy. These tumors demonstrated histological features of adenomatous change and nuclear β-catenin localization, which are characteristic of human neoplasms of the colon and aberrant activation of the Wnt signaling pathway, respectively (Supplementary Figure 1c, Table 1). Mucosal injection of Ad5CMV::Cre similarly induced rapid and efficient tumorigenesis (Supplementary Figure 2a).
Table 1. Efficiency of mucosal injection-based mouse models of colorectal cancer.
Quantification of tumors observed six weeks after mucosal injection. Tumors were identified by colonoscopy and confirmed by necropsy and histology. For all experiments, two injections were performed per mouse. [PGK-Cre: Lentiviral vector with PGK (phosphoglycerate kinase-1) promoter driving expression of Cre recombinase; TU: transforming units; Ad5CMVCre: Adenoviral vector encoding Cre recombinase driven by a cytomegalovirus (CMV) promoter. 4-OHT: 4-hydroxytamoxifen; AKP: Apc and Trp53-null, KrasG12D/+-mutant; NSG: nod SCID gamma]
| Approach | Method | Recipient mouse | Dose per injection (2 injections per mouse) | Number of mice | Number of tumors | Average number of tumors per mouse |
|---|---|---|---|---|---|---|
| Cre-mediated in situ Apc excision | PGK::Cre | Apcfl/fl | 30,000 TU/μl | 16 | 14 | 0.88 |
| Ad5CMV::Cre | Apcfl/fl | 300,000 TU/ μl | 25 | 32 | 1.28 | |
| 4-OH-Tamoxifen | Apcfl/flVillincreER | 100 μM | 35 | 68 | 1.94 | |
| 4-OH-Tamoxifen | Apcfl/flLgr5creER | 100 μM | 10 | 4 | 0.4 | |
| Cre mRNA lipid nanoparticles | Apcfl/fl | 225 μg mRNA/ml | 8 | 7 | 0.88 | |
| CRISPR/Cas9-mediated in situ Apc editing | U6::sgApc-EFS::Cas9-P2A-GFP lentivirus | Wild-type | 1000 TU/μl | 29 | 10 | 0.34 |
| U6::sgApc-CMV::Cre lentivirus | R26LSL-Cas9-eGFP/+ | 10,000 TU/μl | 13 | 12 | 0.92 | |
| U6::sgApc-EFS::turboRFP lentivirus |
R26LSL-Cas9-eGFP/+ VillincreER + Tamoxifen |
10,000 TU/μl | 25 | 25 | 1.0 | |
| CRISPR-Cas9-mediated in situ Trp53 editing | U6::sgTrp53-CMV::Cre lentivirus | Apcfl/fl;R26LSL-Cas9-eGFP/+ | 10,000 TU/μl | 4 | 3 | 0.75 |
| hU6::sgApc-sU6::sgTrp53-EFS::turboRFP lentivirus |
R26LSL-Cas9-eGFP/+ VillincreER + Tamoxifen |
10,000 TU/μl | 4 | 3 | 0.75 | |
| Mouse tumor orthotopic transplantation | AKP mouse CRC cell line | C57BL/6 | 10,000 cells / injection | 6 | 11 | 1.83 |
| sgApc wild-type colon organoids | NSG | ~40 organoids / injection | 25 | 36 | 1.44 | |
| sgApc C57BL/6 wild-type intestinal organoids | C57BL/6 | ~40 organoids / injection | 12 | 10 | 0.83 | |
| AKP colon organoids | NSG | ~40 organoids / injection | 10 | 15 primary 3 liver metastases (week 12) |
1.5 0.33 liver metastasis (week 12) |
|
| AKP colon C57BL/6 organoids | C57BL/6 | ~40 organoids / injection | 20 | 17 | 0.85 | |
| sgTrp53, Apc-null colon organoids | NSG | ~40 organoids / injection | 4 | 5 | 1.25 | |
| Human tumor orthotopic transplantation | Human CRC cell line (LS174 and HT29) | NSG | 10,000 cells / injection | 10 | 18 | 1.8 |
| Patient-derived primary CRC | NSG | 10,000 cells / injection | 10 | 13 | 1.3 | |
| Patient-derived primary CRC organoid | NSG | ~150 organoids / injection | 26 | 24 primary 8 liver metastasis (week 8+12) |
0.92 0.33 liver metastasis (week 8+12) |
|
| Patient-derived MSI-H (Patient B) primary CRC organoid | Humanized NSG | ~150 organoids / injection | 4 | 4 | 1 |
We next generated inducible tumor models using the tamoxifen-dependent Cre recombinase, CreER. CreER driven by the Villin (epithelial-specific) and Lgr5 (intestinal stem cell-specific) promoters has been used to induce intestinal tumors24–26. We generated Rosa26LSL-tdTomato/+;VillinCreER, Apcfl/fl;VillinCreER and Apcfl/fl;Lgr5eGFP-CreER/+ mice and activated CreER locally by mucosal delivery of 4-hydroxytamoxifen (4-OHT). We observed epithelial-specific recombination in Rosa26LSL-tdTomato/+;VillinCreER mice and efficient tumor formation in Apcfl/fl;VillinCreER mice (Supplementary Figure 2b, 2c). These tumors were biopsied for histology (Supplementary Figure 2c). The tumor initiation rate was lower in Apcfl/fl;Lgr5eGFP-CreER/+ mice, most likely due to mosaic expression of the Lgr5 allele in the colon (Supplementary Figure 2d, Table 1)27. We also efficiently induced tumorigenesis in Apcfl/fl mice with stable delivery of Cre mRNA in lipid nanoparticles (Supplementary Figure 3a–e, Table 1). Together, these findings establish CRC mouse models based on Cre-mediated excision of floxed Apc alleles in which tumorigenesis is restricted to the distal colon.
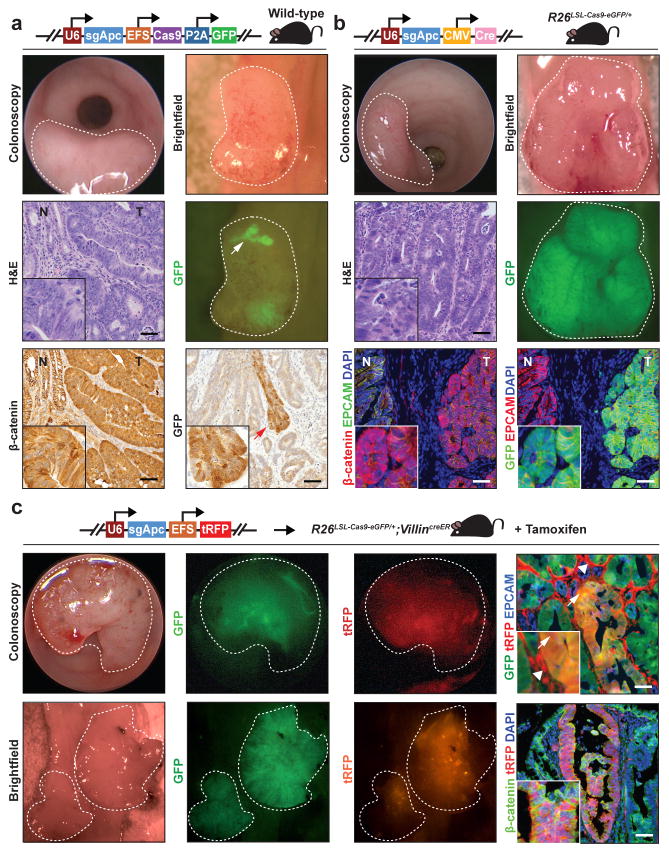
We then sought to harness the mucosal injection approach to model CRC using CRISPR–Cas9-based editing of Apc. We cloned a previously validated short guide RNA (sgRNA) targeting exon 16 of the murine Apc gene28 into a lentiviral vector containing Cas9 and GFP (U6::sgApc-EFS::Cas9-P2A-GFP)29. Mucosal delivery of this virus into wild-type mice resulted in tumors that expressed GFP, indicating stable lentivirus integration. GFP expression was lost in large regions of these adenomas, implicating silencing of lentiviral transgene expression (Figure 1a). In this system, tumorigenesis occurred in 34% of mice receiving two injections, most likely due to relatively low viral titers (approximately 1000 TU/μl) secondary to the large size of the lentiviral genome (~8.1 kb) necessitated by the inclusion of Cas9 (Table 1).
Figure 1. CRISPR-Cas9-based in situ Apc editing in the colon epithelium induces adenoma formation.
(a) Tumors in wild-type mice following mucosal injection with 1,000 transforming units (TU) per μl of U6::sgApc-EFS::Cas9-P2A-GFP lentivirus. Tumors are indicated with white light colonoscopy, brightfield necropsy, GFP fluorescence necropsy, Hematoxylin and eosin (H&E) staining, and β-catenin immunohistochemistry. Note patchy GFP expression in tumors (arrow). (b) Tumors in Rosa26LSL-Cas9-eGFP/+ mice after mucosal delivery of a lentiviruses encoding an sgRNA against Apc and Cre recombinase (U6::sgApc-CMV::Cre, 10,000 TU/μl) (white light colonoscopy, brightfield necropsy, and GFP fluorescence necropsy; H&E immunohistochemistry and GFP/β-catenin immunofluorescence). GFP tumor fluorescence indicates Cre-induced expression of Cas9 and eGFP from the Rosa26 locus. (c) Tumorigenesis in Rosa26LSL-Cas9-eGFP/+;VillinCreER mice treated with tamoxifen and then injected with lentiviruses encoding sgApc and turboRFP (U6::sgApc-EFS::turboRFP, 10,000 TU/μl) into the colon mucosa (white light/GFP/turboRFP fluorescence colonoscopy and brightfield/GFP/turboRFP fluorescence necropsy; GFP/turboRFP immunofluorescence). Arrowheads indicate turboRFP expression in stromal cells. Arrows point to GFP/turboRFP dual-positive tumor cells. Histology images are 20X and insets are 60X (Scale bar: 200 μm). Dotted lines indicate tumors. (tRFP: turboRFP; R26: Rosa26; N: normal; T: tumor).
Massively parallel sequencing of the genomic region flanking the sgRNA target site revealed clonal CRISPR–Cas9-mediated frameshift insertions or deletions in Apc. Notably, we observed distinct Apc-inactivating mutations arising in equal biallelic proportions, which suggests that these tumors originated from two cells (Supplementary Figure 4a). Thus, we demonstrate the application of CRISPR–Cas9 for tracing the clonal origins of cancer, consistent with a recent report of organismal lineage tracing using CRISPR–Cas9.30 This approach is useful for determining the types of mutations that are most potent in transforming cells and thereby confer growth advantage within a multiclonal tumor.
To increase viral titer, we generated a lentivirus containing the Apc sgRNA without Cas9 (U6::sgApc-CMV::Cre, or pUSCC; genome size ~4.4 kb), which produced viral titers of approximately 10,000 TU/μl. Mucosal injection of pUSCC-sgApc into Rosa26LSL-Cas9-eGFP/+ mice31 resulted in tumor formation in 92% of mice (Figure 1b, Table 1). Having shown that mucosal injection of lentivirus results in widespread stromal cell infection (Supplementary Figure 1c), we sought to restrict CRISPR–Cas9 gene editing to colon epithelial cells for cancer modeling. We administered tamoxifen to Rosa26LSL-Cas9-eGFP/+;VillinCreER mice to express Cas9 specifically in intestinal epithelial cells. Mucosal delivery of a U6::sgApc-EFS::turboRFP (pUSET; genome size ~3.8 kb) lentivirus resulted in efficient tumorigenesis characterized by infection of stromal and epithelial cells and CRISPR-mediated Apc editing only in Cas9-positive epithelial cells (Figure 1c Table 1). Consistent with a 10-fold increase in viral titer compared to the U6::sgApc-EFS::Cas9-P2A-GFP lentivirus, we detected multiple frameshifting Apc mutations present in similar proportions, indicating polyclonal tumors that arose from >10 cells of origin (Supplementary Figure 4b). U6::sgApc-EFS::turboRFP tumors that progressed for one year demonstrated high grade dysplasia on histology but no local invasion or liver metastasis (Supplementary Figure 5a, 5b, and data not shown). In comparison to six-week-old tumors, one-year-old U6::sgApc-EFS::turboRFP tumors exhibited dominance of 1–2 Apc frameshifting mutations. These mutations most likely provided a growth advantage to the initiating cells (Supplementary Figure 4b).
To demonstrate the application of our in vivo gene editing system for modeling cancer-associated genes, we performed mucosal editing of the tumor suppressor Trp53 alone (i.e., pUSCC-sgTrp53 into Apcfl/flRosa26LSL-Cas9-eGFP/+ mice) or in combination with Apc (i.e., hU6::sgApc-sU6::sgTrp53-EFS::turboRFP into Rosa26LSL-Cas9-eGFP/+;VillinCreER mice treated with tamoxifen) (Supplementary Figure 5c, 5d, Table 1). Massively parallel sequencing of the genomic region flanking the sgTrp53 target site revealed multiple frameshifting mutations in these tumors (Supplementary Figure 6a, 6b). Together, these studies demonstrate the utility and multiplexability of in vivo somatic CRISPR–Cas9 editing for CRC modeling and assessment of gene function in mice without germline cancer predisposing mutations.
We sought to use our mucosal injection approach to develop an in vivo model of intestinal organoid function in which cultured intestinal organoids are grown in the native colon environment of a host mouse. We derived intestinal and colonic organoids from Rosa26LSL-tdTomato/+;VillinCreER mice treated with tamoxifen and orthotopically transplanted these tdTomato+ organoids into recipient mice by mucosal injection. The engrafted organoids were visualized in vivo by fluorescence colonoscopy (Supplementary Figure 7a). To demonstrate that the engrafted organoids are functional, we: 1) labelled EdU+ proliferating organoid cells; and 2) administered tamoxifen to mice that received Apcfl/fl;VillinCreER intestinal organoids, and observed tumor formation (Supplementary Figure 7a, 7b). Unlike reports of intestinal organoid transplantation by rectal enema into mice32,33, our approach does not require colitis or mechanical injury.
We exploited our mucosal injection technique to model CRC by orthotopically transplanting cancer cells. Syngeneic transplantation of an AKP murine CRC cell line derived from a genetically engineered mouse tumor15 resulted in reproducible engraftment and invasive tumor formation (Supplementary Figure 8a, Table 1). However, these tumors did not reproduce the glandular architecture of human colon adenocarcinoma.
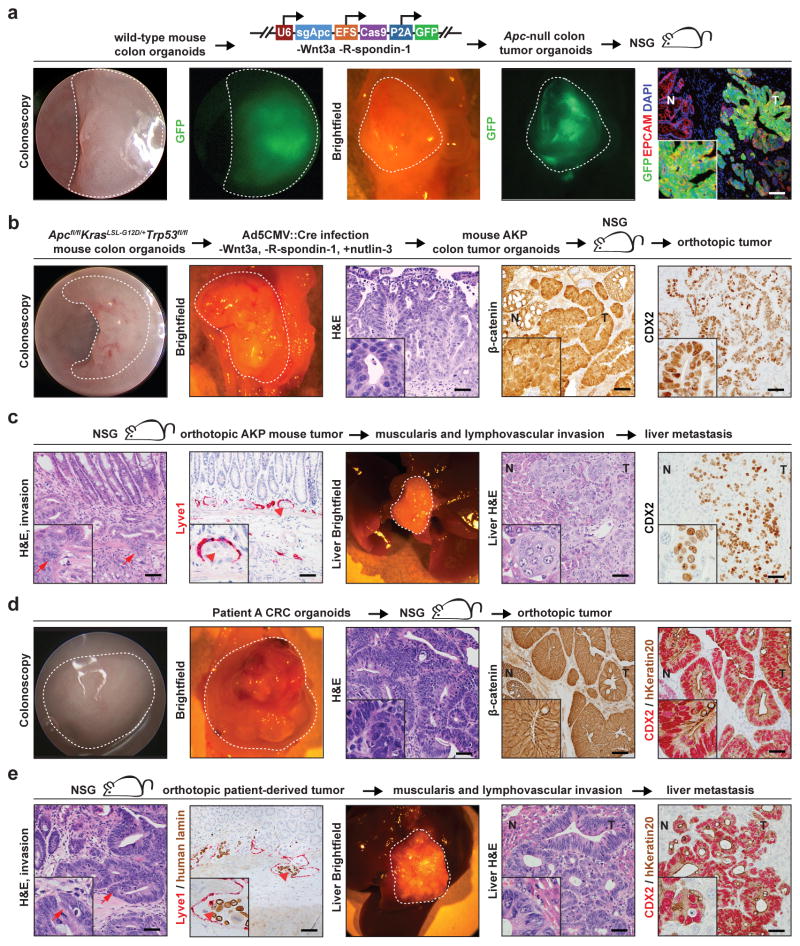
We hypothesized that Apc-deficient intestinal organoids would engraft and form tumors in the distal colon following orthotopic transplantation. Intestinal and colon organoids were infected with U6::sgApc-EFS::Cas9-P2A-GFP lentivirus, and then selected for Apc loss by culturing in media without the exogenous Wnt ligands Wnt3a and R-spondin-1 (Supplementary Figure 8b). These organoids exhibited CRISPR–Cas9-mediated editing at the sgApc target locus and activation of the Wnt signaling pathway by quantitative real-time polymerase chain reaction (qRT-PCR) for Wnt target genes (Supplementary Figure 8c–d, Supplementary Figure 9a, Supplementary Table 1). Orthotopic xenograft and syngeneic transplantation of CRISPR–Cas9-edited Apc-null intestinal organoids into immunodeficient mice and C57BL/6 mice, respectively, resulted in adenomas that extended to the epithelial surface with nuclear β-catenin localization (Figure 2a, Supplementary Figure 8e, Table 1). Analysis of mutations in these tumors at the sgApc binding site revealed substantial intratumoral and intertumoral heterogeneity (Supplementary Figure 9b, 8c). We did not find local invasion or liver metastasis in Apc-null orthotopic tumors in imunodeficient (N=16) and syngeneic recipients (N=20) that progressed for 24 weeks (data not shown). No tumors formed from transplantation of Apc-null intestinal organoids into the mouse flanks of syngeneic mice, which suggests that the colon mucosa is a more permissive environment for intestinal organoid engraftment than the subcutaneous space (N=5, data not shown). Our mucosal injection system complements the enema model of organoid orthotopic transplantation that is described by O’Rourke et al. in this issue.
Figure 2. Orthotopic transplantation models of mouse and patient-derived colorectal cancer.
(a) Tumors in the distal colons of NSG mice following orthotopic transplantation of wild-type colon organoids infected with U6::sgApc-EFS::Cas9-P2A-GFP lentivirus. Tumors are visualized with white light/GFP fluorescence colonoscopy, white light/GFP fluorescence necropsy, and GFP immunofluorescence. (b) Tumors induced in NSG mice by orthotopic transplantation of ApcΔ/ Δ, KrasG12D/+, Trp53Δ/ Δ (AKP) colon organoids. Tumors are imaged with colonoscopy, necropsy, hematoxylin and eosin (H&E) staining, β-catenin immunohistochemistry, and CDX2 immunohistochemistry. (c) Local invasion and liver metastases of engrafted AKP colon organoid tumors, as indicated H&E and LYVE1 immunohistochemistry of primary colon tumors, necropsy, and CDX2 immunohistochemistry of liver metastases. Arrows indicate invasion of the muscularis propria and arrowheads demonstrate tumor invasion of a LYVE1-positive lymphatic vessel. (d) Tumors in NSG mice following orthotopic transplantation of patient-derived CRC organoids. Tumors are demonstrated with colonoscopy, necropsy, H&E staining, β-catenin immunohistochemistry, and CDX2/human Keratin20 immunohistochemistry. (e) Local invasion and liver metastases of patient-derived organoid orthotopic tumors are demonstrated by H&E staining, LYVE1 immunohistochemistry, liver necropsy, and CDX2/human Keratin20 immunohistochemistry. Arrows denote invasion of the muscularis and arrowheads indicate tumor invasion of a LYVE1 positive vessel. Histology images are 20X and insets are 60X (Scale bar: 200 μm). Dotted lines indicate tumors. (NSG: nod SCID gamma; T: tumor; N: normal colon; hKeratin20: human Keratin 20).
We modeled more advanced CRCs by infecting Apcfl/fl;KrasLSL-G12D/+;Trp53fl/fl colon organoids with Ad5CMV::Cre, then selecting in media that contained nutlin-3 and lacked Wnt pathway agonists to generate AKP tumor organoids. These organoids developed invasive tumors with a desmoplastic stromal reaction after orthotopic, syngeneic transplantation, a cardinal feature of invasive human CRC (Supplementary Figure 8f, Table 1). 12 weeks following orthotopic engraftment into NSG mice, AKP organoids invaded the muscularis propria, local vasculature, and metastasized to the liver in 33% of recipient mice (Figure 2b–c, Table 1). All mice with liver metastases exhibited primary colon tumors with invasion of the muscularis propria. These results indicate that orthotopic transplantation of murine tumor organoids can be used to model the entire spectrum of human CRC, including distant-organ metastasis.
Finally, we asked whether in vitro CRISPR–Cas9-based editing of organoids, followed by orthotopic transplantation, can be applied to assess gene function in vivo. We infected colon organoids derived from Apcfl/flRosa26LSL-Cas9-eGFP/+ mice with U6::sgTrp53-CMV::Cre lentivirus, then selected in media without Wnt pathway agonists, followed by selection for Trp53-null organoids with nutlin-3. We then orthotopically engrafted Trp53-null tumor organoids into NSG mice to model P53 mutant CRC (Supplementary Figure 8g, Supplementary Figure 10a, 10b, Table 1).
We subsequently aimed to develop patient-derived orthotopic mouse models of CRC. Engraftment of human CRC cell lines efficiently formed tumors that extended to the surface of the epithelium and invaded the muscularis propria, but did not reflect the histology of human CRC (Supplementary Figure 11a, 11b, Table 1). In contrast, patient-derived orthotopic organoid xenografts produced tumors that accurately recapitulated the epithelial and stromal histology of the original cancer. In addition, patient-derived organoid transplants formed invasive colon tumors that metastasized to the liver in 25% of mice with primary tumors at 8 weeks after transplantation and in 45% of mice at 12 weeks. (Figure 2d, 2e, Supplementary Figure 12a, 12b, 12d, 12e, Table 1, Table 2). Patient-derived CRC organoids transplanted into the mouse flank formed histologically similar tumors but did not metastasize (N=5 tumors; Supplementary Figure 12c and data not shown). Engraftment of patient-derived orthotopic xenografts, in which CRC tissue was digested and directly transplanted into the distal colon without exposure to tissue culture conditions, also engendered tumors that exhibited histological features of human CRC (Supplementary Figure 11c, Table 1). These findings demonstrate that orthotopic transplantation of patient-derived CRC organoids robustly models primary and metastatic human CRC.
Table 2. Clinical characteristics of patient-derived organoids and outcomes of orthotopic transplantation experiments.
All patient-derived organoid transplantation studies were performed in NSG mice. Tumors were identified by colonoscopy and confirmed by necropsy and histology. For all experiments, two injections were performed per mouse. (T: Tumor; N: Node; M: Metastasis; MSS: microsatellite stable; MSI-H: microsatellite instability-high).
| Patient | Age | Sex | Location / pathology | Stage | Mutations | Primary tumors / mice | Liver metastases / primary tumors | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Week 4 | Week 8 | Week 12 | Week 24 | |||||||
| A | 61 | Female | Moderately differentiated liver metastasis from rectal adenocarcinoma | T4 M1 |
MSS. Mutations in KRAS, TP53, PTCH1 | 17/18 | 0/7 | 1/3 | 2/4 | 2/3 |
| B | 68 | Female | Right colon adenocarcinoma with peritumoral lymphocytic infiltrate | Tis N0 M0 |
MSI-H, loss of MSH6 nuclear expression on IHC. KRAS wild-type. Mutations in TP53 and PIK3CA | 12/13 | 0/3 | 1/5 | 2/4 | - |
| C | 47 | Female | Moderately differentiated peritoneal metastasis from right colon | T4b N1b M1b |
MSS. Mutation in KRAS | 11/11 | 0/4 | 1/4 | 1/3 | - |
| All patients (%) | - | - | - | - | - | 40/42 (95%) | 0/14 (0%) | 3/12 (25% | 5/11 (45%) | 2/3 (67%) |
Microsatellite unstable CRC is associated with prominent lymphocytic infiltration and improved survival with anti-programmed death 1 (PD-1) immune checkpoint inhibition.34 However, xenograft models of patient-derived CRC lack a human immune system and therefore have limited utility for studying tumor immunology or immune checkpoint blockade. We derived organoids from a microsatellite instability-high (MSI-H) CRC from a patient with Lynch Syndrome (Patient B; see Table 2). We then orthotopically transplanted the MSI-H organoids into NSG mice with a reconstituted human immune system, which elicited a human lymphocytic infiltrate that is similar to what is observed in MSI-H patient tumors. (Supplementary Figure 12f).35
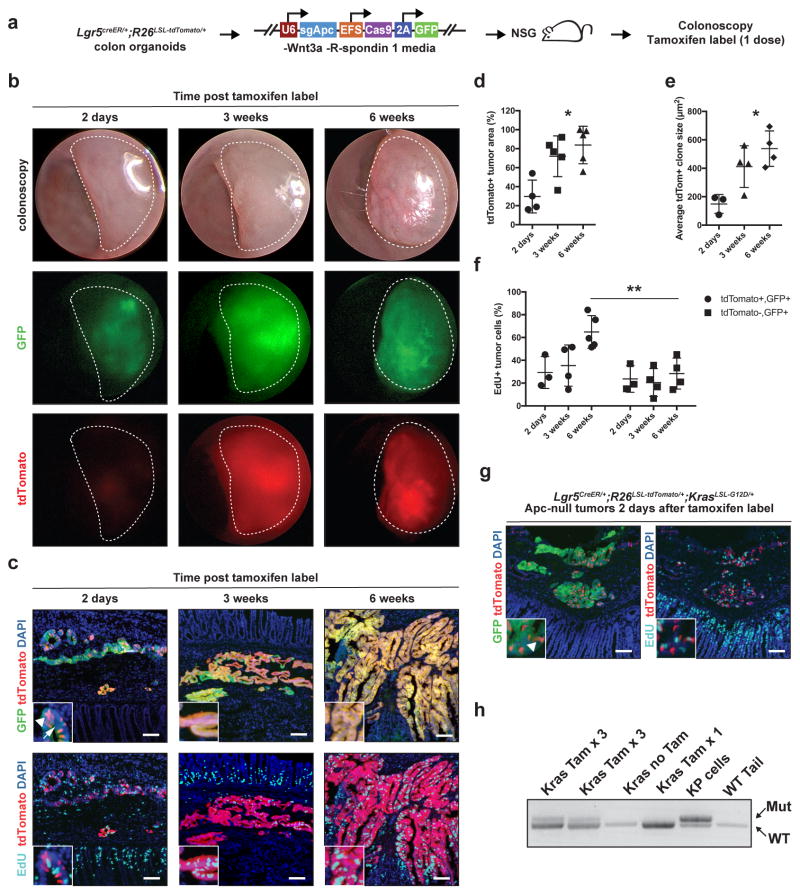
Lgr5 (leucine-rich repeat–containing heterotrimeric guanine nucleotide–binding protein–coupled receptor 5) marks a subpopulation of small intestinal adenoma stem cells that generate new adenoma cells.26 However, the mouse models previously used for Lgr5 cell labelling and retracing do not model colon adenomas and do not permit gene mutation in established adenomas. We used our orthotopic transplantation system to overcome these limitations and lineage trace Lgr5+ cells in established colon adenomas. We generated Lgr5CreER/+;Rosa26LSL-tdTomato/+ mice, derived colon organoids, inactivated Apc by in vitro lentiviral infection (i.e., U6::sgApc-EFS::Cas9-P2A-GFP), and orthotopically engrafted these tumor organoids into NSG mice to generate tumors in vivo that were visualized by colonoscopy two weeks post-injection. Tumor-bearing mice were then administered a tamoxifen pulse to label Lgr5+ cells and their progeny with tdTomato (Figure 3a). Tumors were visualized with fluorescence colonoscopy and immunofluorescence 2 days, 3 weeks, and 6 weeks after labeling (Figure 3b, 3c). Lgr5 cell-derived populations increased in proportion to the total tumor area and in average clone size at 3 and 6 weeks post labelling compared to labelling at 2 days (Figure 3d–e). Notably, tdTomato+ areas contained more proliferating EdU+ cells compared to tdTomato− areas of the tumors, indicating that clones derived from Lgr5+ cells harbor increased proliferative potential (Figure 3f). Finally, we activated oncogenic Kras in Lgr5+ tumor cells by administrating tamoxifen to Apc-null Lgr5CreER/+;Rosa26LSL-tdTomato/+;KrasLSL-G12D/+ orthotopic tumors (Figure 3g–h). These results reveal tumor stem cell activity for Lgr5 tumor cells in colon adenomas and demonstrate the application of our orthotopic transplantation model for sequential mutagenesis in established colon tumors.
Figure 3. Lgr5 cell lineage tracing and sequential mutagenesis in established orthotopic colon adenomas.
(a) Colon organoids derived from Lgr5CreER/+;Rosa26LSL-tdTomato/+ mice were subjected to CRISPR-Cas9-based Apc editing with U6::sgApc-EFS::Cas9-P2A-GFP infection, and then orthotopically transplanted into NSG mice to generate Apc-null in vivo tumors. Tumor-bearing mice received one dose of tamoxifen to label Lgr5+ cells and their progeny with tdTomato and were evaluated 2 days, 3 weeks, or 6 weeks later. Proliferating cells were marked with a 5-ethynyl-2′-deoxyuridine (EdU) pulse 4 hours before sacrifice. (b) In vivo tumor imaging by white light, GFP fluorescence, and tdTomato fluorescence colonoscopy (dotted lines) at 2 days, 3 and 6 weeks after cell labeling. (c) GFP, tdTomato, and EdU immunofluorescence images of orthotopic tumors. Arrowhead indicates cell labeling with tdTomato at 2 days post label (white arrowhead). Arrow points to GFP-negative tumor areas that suggest mosaic lentiviral silencing. Immunofluorescence images were analyzed for: (d) tdTomato+ tumor area relative to total GFP+ tumor area; (e) average tdTomato+ clone size (i.e., total tdTomato+ area / total clone number); and (f) proportion of EdU+ proliferating tdTomato+, GFP+ tumor cells vs. EdU+ proliferating tdTomato−, GFP+ tumor cells at 6 weeks post labelling. (N=3 or 4 tumors for each group and time point). (g) Clonal activation of tdTomato+ (arrowhead) 2 days after tamoxifen administration to mice bearing orthotopic Apc-null Lgr5CreER/+;Rosa26LSL-tdTomato/+;KrasLSL-G12D/+ tumors; (h) Analysis of recombination of the lox-stop-lox (LSL) cassette at the mutant KrasLSL-G12D/+ locus in response to tamoxifen-mediated activation of CreER. Tam x 3 indicates tumors where mice received 3 pulses of tamoxifen, and Tam x1 where one pulse was given. Tissues were harvested 48 hours after the last pulse of tamoxifen. Wild-type mouse tail DNA was used as a negative control, and KrasG12D/+;Trp53Δ/ Δ lung adenocarcinoma cell line DNA (KP cells) as a positive control. *P<0.005 (One-way ANOVA), **P=0.01 (Student’s T-test). (R26: Rosa26; NSG: nod SCID gamma; tdTom: tdTomato)
Our in situ epithelial gene editing and orthotopic transplantation systems provide significant advances over standard mouse models of CRC: 1) tumors are located in the appropriate tissue compartment (i.e., the colon) and correct tissue layer (i.e., lamina propria); 2) organoids require only Apc loss for orthotopic engraftment, instead of multiple additional oncogenic mutations that are required for engraftment at other sites21,22; 3) tumors form within a few weeks; 4) tumors are seen in almost all experimental mice; 5) tumors are longitudinally monitored with colonoscopy; 6) tumors with advanced mutations reproduce key pathological features of human CRC, including progression from primary cancer, invasion of the muscularis propria, and liver metastasis 36,5,37; 7) customized viral vectors reduce the cost and time required to functionally interrogate cancer-associated genes; 8) tumors are quickly induced with defined CRISPR–Cas9-based genetic alterations in vivo or in organoids without the time-consuming need to generate mutations in the germline; and 9) orthotopic transplantation of Apc-edited organoids permits lineage tracing and sequential mutagenesis in established adenomas.
An important goal of cancer modeling is to reproduce primary and metastatic disease in vivo for preclinical research and for clinical applications. Patient-derived CRC organoids have been shown in vitro to recapitulate molecular, genetic, and pathological features of the original tumors, but to date have been engrafted only into the kidney capsule and flank. Organoid-based models of metastatic CRC are limited to seeding of distal organs from the kidney capsule or spleen38,39. We demonstrate the use of patient-derived organoids to model tumor formation in the native colon environment with tumor and stromal histology that accurately reflects the patient’s disease. Finally, we apply our orthotopic transplantation system to model patient-derived primary cancer, local tumor invasion, and liver metastasis.
Methods
Mice
Mice were housed at the animal facility at the Koch Institute for Integrative Cancer Research at MIT. All animal studies described in this study were approved by the MIT Institutional Animal Care and Use Committee. Apcfl/fl (ref. 40), KrasLSL-G12D/+ (ref. 41), VillinCreER (ref. 42), Lgr5eGFP-CreE (ref. 27), Lgr5CreER (ref. 43), and Rosa26LSL-Cas9-eGFP (ref. 31) mice have been described and were maintained on pure C57BL/6 or mixed C57BL/6J × 129SvJ backgrounds. Rosa26LSL-tdTomato mice44 were maintained on a mixed C57BL/6J × 129SvJ genetic background. Orthotopic transplantation experiments with C57BL/6 organoids were performed into syngeneic C57BL/6 or NOD scid gamma (NSG)45 recipient mice. Mixed background mouse organoids and human organoids engraftments were performed with NSG recipient mice. Humanized mice were developed as previously described.45 Approximately equal numbers of male and female mice of 6–10 weeks age were used for all experiments.
Generation of lentiviral vectors and sgRNA cloning
The pL-CRISPR-EFS::GFP vector was obtained from Addgene (#57818)29. The pSECC vector has been previously described and is available from Addgene (#60820)19. The U6::sgRNA-CMV::Cre (pUSCC) or U6::sgRNA-EFS::turboRFP (pUSET) lentiviral vectors were constructed by assembling four parts with overlapping DNA ends using modular Gibson assembly of lentiviruses, as previously described46. Briefly, a 2.2kb part (corresponding to the U6-Filler fragment from pSECC), a 0.3kb part (corresponding to the EFS promoter from pSECC), a 0.5kb part [corresponding to the CMV promoter from CMV-GFP, for pUSCC (Addgene #68485)46], a 0.7kb part [corresponding to turboRFP in pTRIPZ (Dharmacon, catalogue # RHS4740), for pUSET] or a 1kb part (corresponding to Cre recombinase from pSECC, for pUSCC), and a 5.7kb lentiviral backbone were assembled using Gibson assembly following the manufacturer’s protocol (New England Biolabs Gibson Assembly Master Mix; catalogue number E2611S). For sgRNA cloning, the pUSCC, pUSET and U6-sgApc-EFS-Cas9-2A-GFP vectors were digested with BsmBI and ligated with a BsmBI-compatible annealed sgRNA oligo targeting exon 16 in Apc (target sequence GTCTGCCATCCCTTCACGTTAGG, PAM sequence underlined), as previously reported.28 The pUSCC vector was digested and ligated with a previously described sgRNA targeting exon 7 in Trp53 (target sequence GTGTAATAGCTCCTGCATGGGGG, PAM sequence underlined).47 The paired guide hU6-sgApc-sU6-sgTrp53-EFS-turboRFP vector was cloned using the above sgRNAs using a previously described system.48
Lentivirus production
Lentivirus was produced by co-transfection of 293T cells with lentiviral backbone constructs and packaging vectors (pSpax2 and pMD2.G) using TransIT-LT1 (Mirus Bio). Supernatant was collected 48 and 72 hours post-transfection, concentrated by ultracentrifugation at 25,000 rpm for two hours and resuspended in an appropriate volume of OptiMEM (Invitrogen, catalogue # 31985-070). Viral titer was calculated by serial infection of 293T cells (for lentiviruses containing either GFP or turboRFP) or GreenGo Cre reporter cells (for lentiviruses containing Cre).19
Murine intestinal organoid culture and lentiviral infection
As previously reported23 and briefly summarized here, small intestines and colons were removed, washed with cold PBS−/−, opened laterally and cut into 3–5 mm fragments. Pieces were washed multiple times with cold PBS−/− until clean, washed 2–3 with PBS−/−/EDTA (10mM), and incubated on ice for 90–120 minutes while mixing at 30-minute intervals. Crypts were then mechanically separated from the connective tissue by shaking, and filtered through a 70-μm mesh into a 50-ml conical tube to remove villus material (for small intestine) and tissue fragments. Crypts were then resuspended in 50% L-WRN conditioned media as previously described,49 henceforth referred to as conditioned media (derived from L-WRN cells, a kind gift from Thaddeus Stappenbeck, Washington University) and manually counted. Crypts were embedded in growth factor-reduced Matrigel™ (Corning, catalogue # 356231), diluted 3:4 in conditioned media into 24 well plates (Olympus, catalogue # 25-107) at a density of approximately 750 crypts in 75 μl total volume per well, and incubated with conditioned media supplemented with 10 μM of the p160 ROCK inhibitor Y-27632 dihydrochloride monohydrate (APExBIO, catalogue # A3008) to prevent cell death by anoikis.
Murine intestinal organoids were infected with lentivirus based on a protocol previously described by the Hans Clevers lab for organoid retroviral infection.50 Briefly, 2–3 days following culture of intestinal crypts, media was changed for culture media plus 10 mM nicotinamide (Sigma Aldrich, catalogue # N3376). At this point, organoids were cystic or round in appearance, which indicates the presence of healthy stem cells. The following day, organoids were disrupted mechanically by pipetting up and down 30 to 50 times with a 1000 μl pipette. At this stage, the organoids were ideally small clumps of < 10 cells. If mechanical dissociation was not sufficient, then organoids were gently dissociated enzymatically with 1X TrypLE Express (Life Technologies, catalogue # 12604-021) at 37 degrees C for 1–5 minutes. Organoids were then resuspended in a 24 well plate with culture media supplemented with 10 μM Y27632, 10 mM nicotinamide, and 8 μg/ml Polybrene (Sigma Aldrich, catalogue # TR-1003). The plate was centrifuged for 60 minutes at 600 g at 32 degrees C (spinoculation), and then incubated at 37 degrees C for 6 hours. Infected organoids were then embedded in Matrigel and cultured with culture media plus 10 μM Y27632 and 10 mM nicotinamide. For sgApc CRISPR–Cas9 viral infections, organoids were maintained in conditioned media for seven days to permit Cas9-mediated editing of Apc, then in media without Wnt3a or R-spondin-1 [Advanced DMEM/F12 (Life Technologies, catalogue # 12634-028) supplemented with 1X N2 (Life Technologies, catalogue # 17502-048) and 1X B27 (Life Technologies, catalogue # 17504-044)] for selection of Apc-deficient organoids.
AKP colon organoids were generated by culturing colon organoids from Apcfl/fl;KrasLSL-G12D/+;Trp53fl/fl mice and infecting with Ad5CMV::Cre. CRISPR-Trp53 colon organoids were created by culturing organoids from Apcfl/fl;R26LSL-Cas9-eGFP/+ mice and infecting with U6::sgTrp53-CMV::Cre lentivirus. AKP and ApcΔ/Δ, p53-null organoids were selected in media without Wnt3a or R-spondin-1 supplemented with 10 μM nutlin-3 (Cayman Chemical, catalogue # 10004372).
Murine colonoscopy and mucosal injection
Optical and fluorescence colonoscopy was performed using the Image 1 H3-Z Spies HD Camera System (part TH100), Image 1 HUB CCU (parts TC200, TC300), 175 Watt D-Light Cold Light Source (part 20133701-1), AIDA HD capture system, and 0″ Hopkins Telescope (part 64301AA), and fluorescent filters in the tdTomato (emission 554 nm) and GFP channels (emission 509 nm) (all from Karl Storz).
For mucosal injections, Ad5CMV::Cre, 4-OHT (Calbiochem, catalogue #579002) or lentivirus were resuspended in OptiMEM, then delivered to the colonic lamina propria of C57BL/6 recipient mice (ideally 6–10 weeks old) by optical colonoscopy using a custom injection needle (Hamilton Inc., 33 gauge, small Hub RN NDL, 16 inches long, point 4, 45-degree bevel, like part number 7803-05), a syringe (Hamilton Inc., part number 7656-01), a transfer needle (Hamilton Inc., part number 7770-02), and a colonoscope with integrated working channel (Richard Wolf 1.9mm/9.5 French pediatric urethroscope, part number 8626.431). 2–3 injections containing 50–100 μl of media were performed per mouse. For orthotopic organoid transplantations, Apc-null organoids were gently mechanically dissociated, resuspended in 90% minimal media (Advanced DMEM plus N2 and B27) and 10% Matrigel, and then transplanted into recipient mice. Mice underwent colonoscopy 4–8 weeks following lentiviral injection or organoid transplantation to assess tumor formation. Colonoscopy videos and images were saved for offline analysis. Tumor size was quantified as previously described using ImageJ.51 Following sacrifice, the distal colons were excised and fixed in 10% formalin, sectioned and examined by hematoxylin and eosin staining to identify adenomas. Alternatively, tumors were fixed for 1 hour in 4% paraformaldehyde, then overnight in 25% sucrose solution, and frozen for sectioning.
Human colorectal cancer organoid collection and isolation
0.5–1 cm3 portions of fresh CRC surgical specimens were obtained from patients undergoing surgery for CRC at Tufts Medical Center, Massachusetts General Hospital, and Brigham and Women’s Hospital / Dana Farber Cancer Institute. Cancer tissue was selected by the clinical pathologist only if not required for clinical evaluation. Samples were placed in cold PBS and transported to the Koch Institute at MIT. The respective Institutional Review Board committees and the Massachusetts Institute of Technology Committee on the Use of Humans as Experimental Subjects approved the study protocols. Informed consent was obtained from all subjects. Cancer tissues were grown into organoids as previously described,38 with minor modifications. Briefly, tissues were finely minced, then digested with collagenase Type 1 (200 units in 5 ml PBS) on ice for 5 – 10 minutes. After digesting into clumps of cells, the sample was filtered through a 100 μm mesh and seeded into Matrigel in 24-well plates (50–75 μl per well). Following Matrigel polymerization (10 minutes at 37 degrees), 650 μl of human culture media (conditioned media, 1X N2, 1X B27, EGF 40 ng/ml (PeproTech, catalogue # 315-09), 3 μM SB202190 (Sigma Aldrich, catalogue # S7067), 500 nM A83-01 (Tocris, catalogue # 2939), 20 ng/ml Y-27632 dihydrochloride monohydrate, 1 μM N-acetyl-l-cysteine (Sigma-Aldrich), 10 mM nicotinamide, 10 nM human gastrin I (Sigma-Aldrich, catalogue # G9020), and 100 μg/ml Primocin (InvivoGen, catalogue # ant-pm-1). Two days after organoid formation, tumor organoids were selected by changing media to ADMEM supplemented with 1X Penicillin / Streptomycin, 100 ug/ml primocin, 1X N2, and 1X B27.
Immunohistochemistry
As previously described, tissues and organoids were fixed in 10% formalin, paraffin embedded, and sectioned.23,52 Antigen retrieval was performed with Borg Decloaker RTU solution (Biocare Medical) in a pressurized Decloaking Chamber (Biocare Medical) for 3 minutes. The following antibodies were used: mouse monoclonal β-catenin (1:200, BD Biosciences 610154), rabbit monoclonal CDX2 (1:1250, Abcam ab76541), rabbit monoclonal human specific Cytokeratin 20 (1:500, Abcam, ab76126), rabbit polyclonal Lyve 1 (1:200, Abcam, ab14917), rabbit monoclonal Anti-CD3 (1:200, Abcam ab16669), CD31 (1:50, Abcam, ab28364), and the human specific nuclear envelope marker Lamin A+C (1:2500, Abcam, ab108595). Biotin-conjugated secondary donkey anti-rabbit or anti-rat antibodies were used from Jackson ImmunoResearch. The Vectastain Elite ABC immunoperoxidase detection kit (Vector Labs PK-6101) followed by Dako Liquid DAB+ Substrate (Dako) was used for visualization. Hematoxylin and eosin (H&E) and trichrome stains were performed according to standard procedures.
Immunofluorescence
Immunofluorescence on cryosections was performed as previously described53. Briefly, 7 μm cryosections of intestines were air-dried, fixed with cold acetone, washed with PBS and blocked with Donkey Immunomix (5% normal donkey serum, 0.2% bovine serum albumin, 0.05% sodium azide, and 0.3% Triton-X100 in PBS). The following primary antibodies were used for immunostaining of mouse tissues: rabbit β-catenin (1:100, AbCam, catalogue # ab32572), mouse β-catenin (1:100, BD Biosciences, catalogue # 610154), rabbit polyclonal lysozyme (1:250, Thermo Scientific, catalogue # RB-372-A1), rat EpCAM-APC (1:500, Biolegend, catalogue # 17-5791-82). Sections were washed with PBS containing 0.3% Triton-X100 and the primary antibodies were detected with the appropriate Alexa 488, 594 or 647 secondary antibody conjugates (Molecular Probes/Life Technologies). Tissues were post-fixed in 1% paraformaldehyde and mounted in Vectashield with DAPI (VectorLabs, catalogue # H1200). Immunofluorescence images were captured using a Nikon A1R confocal microscope using 10x or 20x air objectives and multichannel scanning in frame mode (pinhole 1.2 Airy units). For identification of proliferating cells, mice harboring orthotopic tumors were injected intraperitoneally with 1 mg of 5-ethynyl-2-deoxyuridine (EdU, Setareh Biotech) 4 hours prior to euthanasia. EdU was detected in cryosections using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermoscientific) according to the manufacturer’s protocol.
Statistics
For analysis of the statistical significance of differences between two groups, we used two-tailed Student’s t-tests based on a normal distribution of the data. All error bars denote standard deviation. All samples represent biological replicates. No samples or animals were excluded from analysis, and sample size estimates were not used. Animals were randomly assigned to groups. Studies were not conducted blind.
Bioluminescence imaging
Colons of wild-type mice were injected with lipid nanoparticles containing firefly luciferase mRNA or firefly luciferase mRNA alone. Rosa26LSL-tdTomato/LSL-Luciferase mice were injected with Ad5CMV::Cre under colonoscopy guidance. Two days later, mice were injected intraperitoneally with 100 mg/kg D-Luciferin, which was allowed to circulate for 10 min, followed by euthanasia and dissection of colons for biouminescence imaging (IVIS, Caliper Life Sciences).
Lipid nanoparticle synthesis
Lipid nanoparticles (LNPs) were synthesized as previously described54. In brief, an ethanol phase containing the lipids and an aqueous phase containing the mRNA were mixed in a microfluidic chip device. The aqueous phase consisted of 300μL of Cre mRNA (1mg/mL in in 10mM TRIS-HCL, from TriLink Biotechnologies, San Diego, CA), 155μL of citrate (100mM, pH 3), and 1097μL of water. The ethanol phase contained the ionizable lipid ckk-E12 (3.0mg, synthesized in our laboratory, as previously described 55, with 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE, 4.1mg, Avanti Polar Lipids, Alabaster, AL), cholesterol (3.3mg, Sigma), and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (C14-PEG 2000, 1.4 mg, Avanti) in ethanol (518μL, Sigma). The resulting LNP solution was dialyzed against PBS in 20,000 MWCO cassette at 20 °C for 2 hours. The size (118nm, 100%) and polydispersity (PDI = 0.128) of the LNP was measured using dynamic light scattering (ZetaPALS, Brookhaven Instruments).
Electron Microscopy
A cryogenic TEM picture was taken of LNPs in a buffered solution on a lacey copper grid coated with a continuous carbon film. The grid was then mounted on a Gatan 626 cryo-holder equipped within in TEM column. The specimen and holder tip were continuously cooled by liquid nitrogen during transfer into the microscope and subsequent imaging. Imaging was performed using a JEOL 2100 FEG microscope using a minimum dose method that was essential to avoiding sample damage under the electron beam. The microscope was operated at 200kV and with a magnification setting of 60,000 for assessing particle size and distribution. All images were recorded on a Gata 2kx2k UltraScan CCD camera.
Massively parallel sequencing
A genomic region containing the sgApc target sequence was amplified using Herculase II Fusion DNA polymerase and gel purified (Forward primer 5′ to 3′: AAGACCAGGAAGCCTTGTGG; Reverse primer 5′ to 3′: GCTTGTGTCTCTGCTTACTCC). Sequencing libraries were prepared from 50ng of PCR product using the Nextera DNA Sample Preparation Kit (Illumina) according to manufacturer’s instructions and sequenced on Illumina MiSeq sequencers to generate 150 bp paired-end reads. The sgTrp53 target sequence was amplified and sequenced using the above methods and the following primers: Forward primer 5′ to 3′: ATTCCCGGCTGCTGCAGGTC; Reverse primer 5′ to 3′: GGCGGGACTCGTGGAACAGAA).
Bioinformatic analysis of target loci
Illumina MiSeq reads (150bp paired-end) were trimmed to 120bp after reviewing base quality profiles, in order to drop lower quality 3′ ends. Traces of Nextera adapters were clipped using the FASTX toolkit (Hannon Lab, CSHL) and pairs with each read greater than 15bp in length were retained. Additionally, read pairs where either read had 50% or more bases below a base quality threshold of Q30 (Sanger) were dropped from subsequent analyses. The reference sequence of the target locus was supplemented with 10bp genomic flanks and was indexed using an enhanced suffix array56. Read ends were anchored in the reference sequence using 10bp terminal segments for a suffix array index lookup to search for exact matches. A sliding window of unit step size and a maximal soft-clip limit of 10bp was used to search for possible anchors at either end of each read. For each read, optimal Smith-Waterman dynamic programming alignment57 was performed between the reduced state space of the read sequence and the corresponding reference sequence spanning the maximally distanced anchor locations. Scoring parameters were selected to allow for sensitive detection of short and long insertions and deletions while allowing for up to four mismatches, and the highest scoring alignment was selected. Read pairs with both reads aligned in the proper orientation were processed to summarize the number of wild-type reads and the location and size of each insertion and deletion event. Overlapping reads within pairs were both required to support the event if they overlapped across the event location. Additionally, mutation events and wild-type reads were summarized within the extents of the sgRNA sequence and PAM site by considering read alignments that had a minimum of 20bp overlap with this region. Mutation calls were translated to genomic coordinates and subsequently annotated using Annovar58. The alignment and post-processing code was implemented in C++ along with library functions from SeqAn59 and SSW60 and utility functions in Perl and R (www.R-project.org). Mutation calls were subjected to manual review using the Integrated Genomics Viewer (IGV)61.
CRC cell lines
Murine CRC cell lines were derived from AKP genetically engineered tumors, as previously described.15 Following mechanical abrasion of the distal colon, 100 μl Ad5CMV::Cre (University of Iowa Viral Vector Core Facility; 1010 TU/μl) was delivered by rectal enema to C57BL/6 Apcfl/fl;KrasLSL-G12D/+;Trp53fl/fl mice during laparotomy to restrict adenoviral delivery to the distal colon and initiate tumorigenesis. Tumors were extracted, finely minced, digested with collagenase Type 1 (200 units in 5 ml PBS) (Worthington, catalogue # LS004196) and grown (DMEM supplemented with 1X penicillin/streptomycin and 10% fetal bovine serum) as immortalized two-dimensional cultures. Cell cultures were then sorted as EpCAM+ single cells by FACS and expanded to derive pure AKP cell lines. LS174 human CRC cells were obtained from ATCC. Both murine AKP and human LS174 cells tested negative for Mycoplasma contamination.
In situ hybridization for human CDX2
Automated ISH assays for human CDX2 mRNA were performed using View- RNA eZL Detection Kit (Affymetrix) on the Bond RX immunohistochemistry and ISH Staining System with BDZ 6.0 software (Leica Biosystems). Formalin-fixed paraffin-embedded (FFPE) tissue sections on slides were processed automatically from deparaffinization, through ISH staining to hematoxylin counterstaining. Briefly, 5 mm-thick sections of formalin-fixed tissue were baked for 1 hour at 60C and placed on the Bond RX for processing. The Bond RX user-selectable settings were as follows: ViewRNA eZ-l Detection 1-plex (Red) protocol; ViewRNA Dewax1 Preparation protocol; View RNA HIER 10 minutes, ER1 (95); ViewRNA Enzyme 2 (10); ViewRNA Probe Hybridization. With these settings, the RNA unmasking conditions for the FFPE tissue consisted of a 10-minute incubation at 95 C in Bond Epitope Retrieval Solution 1 (Leica Biosystems), followed by 10-minute incubation with Proteinase K from the Bond Enzyme Pretreatment Kit at 1:1000 dilution (Leica Biosystems). Human CDX2 Ez probes were diluted as 1:40 and 1:20 respectively in ViewRNA Probe Diluent (Affymetrix). Post run, slides were rinsed with water, air dried for 30 minutes at room temperature and mounted using Dako Ultramount (Dako, Carpinteria, CA), and visualized using a standard bright-field microscope.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (T.J., R.O.H.), NIH (K08 CA198002, J.R.; K99 CA187317, T.T.; R00 AG045144, Ö.H.Y.), Department of Defense (PRCRP Career Development Award CA120198; J.R.), and the V Foundation V Scholar Award (J.R. and Ö.H.Y.), the Sidney Kimmel Scholar Award (Ö.H.Y), the Pew-Stewart Trust Scholar Award (Ö.H.Y), Koch Institute Frontier Research Program through the Kathy and Curt Marble Cancer Research Fund (Ö.H.Y), American Federation of Aging Research (AFAR, Ö.H.Y), the Hope Funds for Cancer Research (T.T.), and by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. We thank Thales Papagiannakopoulos for helpful discussions; Yadira Soto-Feliciano for help and expertise with massively parallel sequencing; Kristi Bedrossian for assistance with human colorectal cancer sample collection at Tufts Medical Center; the Swanson Biotechnology Center at the Koch Institute for technical support, specifically Kathleen Cormier and Charlene Condon at the Hope Babette Tang (1983) Histology Facility; Sven Holder for histology support; and Yun Dong Soo and the Peterson (1957) Nanotechnology Materials Core Facility for assistance with electron microscopy.
Footnotes
Competing Financial Interests Statement: The authors have no competing financial interests
Author Contributions
J.R. and T.T. performed all experiments and participated in their design and interpretation with T.J and Ö.H.Y. J.R. and Ö.H.Y. developed and optimized the colonoscopy mucosal injection technique, with assistance from D.K and P.K. J.R. wrote the paper with support from T.T. and Ö.H.Y. N.M.C. contributed to study design, plasmid design, lentivirus production, and mucosal injections. F.S.R. contributed to plasmid and study design, and performed massively parallel sequencing. M.A., Y.K.P., R.N., R.A., X.L., and A.A. assisted with mucosal injections, mouse and human organoid derivation, molecular biology, and immunohistochemistry. S.R. performed immunohistochemistry experiments. A.R. assisted with humanized mouse experiments. M.A.O. designed and synthesized lipid nanoparticles for mRNA encapsulation. G.E., E.T.S., M.T., A.J. Bass, Y.S., J.Y., L.C., V.D., and L.Z. assisted with human CRC specimen collection. S.B. performed organoid qRT-PCR. A. Bhutkar performed bioinformatics analysis. R.L., J.L., J.C., P.N.T., R.O.H, and T.J. participated in interpretation of results. Ö.H.Y. supervised all aspects of the study.
References
- 1.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 4.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 5.Roper J, Hung KE. Priceless GEMMs: genetically engineered mouse models for colorectal cancer drug development. Trends Pharmacol Sci. 2012;33:449–455. doi: 10.1016/j.tips.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Golovko D, Kedrin D, Yilmaz ÖH, Roper J. Colorectal cancer models for novel drug discovery. Expert Opin Drug Discov. 2015;10:1217–1229. doi: 10.1517/17460441.2015.1079618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinoi T, et al. Mouse Model of Colonic Adenoma-Carcinoma Progression Based on Somatic Apc Inactivation. Cancer Res. 2007;67:9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 8.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue Y, Johnson R, Desmet M, Snyder PW, Fleet JC. Generation of a transgenic mouse for colorectal cancer research with intestinal cre expression limited to the large intestine. Mol Cancer Res MCR. 2010;8:1095–1104. doi: 10.1158/1541-7786.MCR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata H, et al. Rapid Colorectal Adenoma Formation Initiated by Conditional Targeting of the Apc Gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 11.Hung KE, et al. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc Natl Acad Sci U S A. 2010;107:1565–1570. doi: 10.1073/pnas.0908682107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadac JN, et al. Colon Tumors with the Simultaneous Induction of Driver Mutations in APC, KRAS, and PIK3CA Still Progress through the Adenoma-to-carcinoma Sequence. Cancer Prev Res Phila Pa. 2015;8:952–961. doi: 10.1158/1940-6207.CAPR-15-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu XY, Besterman JM, Monosov A, Hoffman RM. Models of human metastatic colon cancer in nude mice orthotopically constructed by using histologically intact patient specimens. Proc Natl Acad Sci U S A. 1991;88:9345–9349. doi: 10.1073/pnas.88.20.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi T, Morotomi M, Nomoto K. A novel mouse model of rectal cancer established by orthotopic implantation of colon cancer cells. Cancer Sci. 2004;95:514–519. doi: 10.1111/j.1349-7006.2004.tb03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin ES, et al. Development of a Colon Cancer GEMM-Derived Orthotopic Transplant Model for Drug Discovery and Validation. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19:2929–2940. doi: 10.1158/1078-0432.CCR-12-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhullar JS, et al. A novel nonoperative orthotopic colorectal cancer murine model using electrocoagulation. J Am Coll Surg. 2011;213 doi: 10.1016/j.jamcollsurg.2011.02.022. 54-60-61. [DOI] [PubMed] [Google Scholar]
- 17.Zigmond E, et al. Utilization of murine colonoscopy for orthotopic implantation of colorectal cancer. PloS One. 2011;6:e28858. doi: 10.1371/journal.pone.0028858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maddalo D, Ventura A. Somatic Engineering of Oncogenic Chromosomal Rearrangements: A Perspective. Cancer Res. 2016;76:4918–4923. doi: 10.1158/0008-5472.CAN-16-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015 doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 22.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015 doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 23.Beyaz S, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto D, Robine S, Jaisser F, El Marjou FE, Louvard D. Regulatory sequences of the mouse villin gene that efficiently drive transgenic expression in immature and differentiated epithelial cells of small and large intestines. J Biol Chem. 1999;274:6476–6482. doi: 10.1074/jbc.274.10.6476. [DOI] [PubMed] [Google Scholar]
- 25.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 26.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 27.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 28.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Heckl D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A, et al. Whole organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016 doi: 10.1126/science.aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yui S, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda M, et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev. 2014;28:1752–1757. doi: 10.1101/gad.245233.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le DT, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, et al. Induction of functional human macrophages from bone marrow promonocytes by M-CSF in humanized mice. J Immunol Baltim Md 1950. 2013;191:3192–3199. doi: 10.4049/jimmunol.1300742. [DOI] [PubMed] [Google Scholar]
- 36.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 37.Roper J, et al. Combination PI3K/MEK inhibition promotes tumor apoptosis and regression in PIK3CA wild-type, KRAS mutant colorectal cancer. Cancer Lett. 2014;347:204–211. doi: 10.1016/j.canlet.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujii M, et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Kuraguchi M, et al. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2006;2:e146. doi: 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 42.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genes N Y N 2000. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 43.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shultz LD, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol Baltim Md 1950. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 46.Akama-Garren EH, et al. A Modular Assembly Platform for Rapid Generation of DNA Constructs. Sci Rep. 2016;6:16836. doi: 10.1038/srep16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malina A, et al. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 2013;27:2602–2614. doi: 10.1101/gad.227132.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidigal JA, Ventura A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat Commun. 2015;6:8083. doi: 10.1038/ncomms9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koo B-K, Sasselli V, Clevers H. Retroviral gene expression control in primary organoid cultures. Curr Protoc Stem Cell Biol. 2013;27(Unit 5A.6) doi: 10.1002/9780470151808.sc05a06s27. [DOI] [PubMed] [Google Scholar]
- 51.Roper J, et al. The Dual PI3K/mTOR Inhibitor NVP-BEZ235 Induces Tumor Regression in a Genetically Engineered Mouse Model of PIK3CA Wild-Type Colorectal Cancer. PloS One. 2011;6:e25132. doi: 10.1371/journal.pone.0025132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yilmaz ÖH, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tammela T, et al. Angiopoietin-1 promotes lymphatic sprouting and hyperplasia. Blood. 2005;105:4642–4648. doi: 10.1182/blood-2004-08-3327. [DOI] [PubMed] [Google Scholar]
- 54.Chen D, et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 55.Dong Y, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abouelhoda MI, Kurtz S, Ohlebusch E. Replacing suffix trees with enhanced suffix arrays. J Discrete Algorithms. 2004;2:53–86. [Google Scholar]
- 57.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 58.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Döring A, Weese D, Rausch T, Reinert K. SeqAn an efficient, generic C++ library for sequence analysis. BMC Bioinformatics. 2008;9:11. doi: 10.1186/1471-2105-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao M, Lee WP, Garrison EP, Marth GT. SSW library: an SIMD Smith-Waterman C/C++ library for use in genomic applications. PloS One. 2013;8:e82138. doi: 10.1371/journal.pone.0082138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.