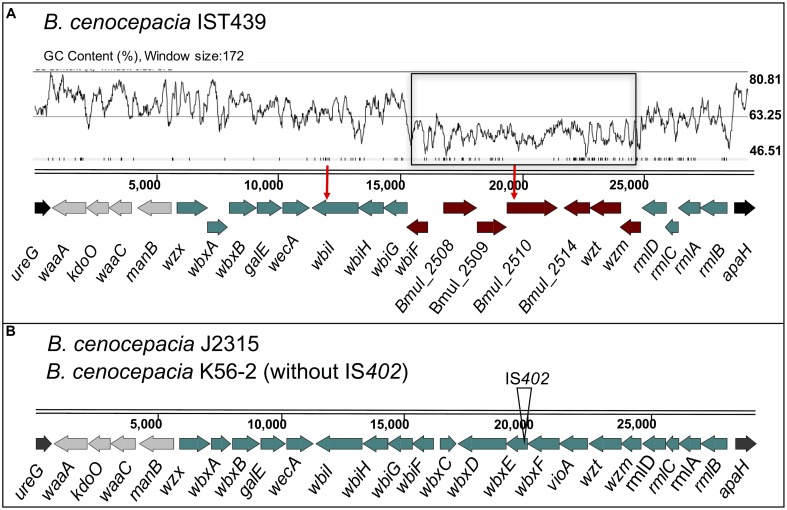
FIGURE 6.
Genetic organization of gene clusters for core-lipid A and OAg biosynthesis in B. cenocepacia IST439 (A) and the reference strains K56-2 and J2315 (B). The flanking genes ureG and apaH are indicated in black and the four genes represented in gray encode proteins putatively involved in lipid A-core biosynthesis. The genes in dark-red correspond to genes present in IST439 without a counterpart in the reference strain J2315 (Supplementary Figure S3), but with a degree of homology to B. multivorans ATCC 17616 (see also Supplementary Figure S4B). The red-vertical arrows above the genes represent the two non-synonymous point mutations in genes bmul_2510 and wbiI of the 10 sequential isolates (comparative genomic analysis to be described elsewhere). A GC content plot is also represented for IST439 [drawn using Artemis (Carver et al., 2008)] above the display line of the sequence, where the genes annotated as B. multivorans are highlighted in a black rectangle. wλ, 3-deoxy-D-manno-octulosonic acid transferase; kdoO, Kdo dioxygenase; waaC, heptosyltransferase I; manB, phosphomannomutase; wzx, OAg exporter; wbxA, glycosyltransferase; wbxB, glycosyltransferase; galE, UDP-glucose epimerase; wecA, UDP-N-acetylglucosamine 1-P transferase; wbiI, nucleotide sugar epimerase-dehydratase; wbiH, UDP-N-acetylglucosamine 1-P transferase; wbiG, nucleotide sugar epimerase-dehydratase; wbiF, glycosyltransferase; bmul_2508, conserved hypothetical protein; Bmul_2509, group 1 glycosyl transferase; Bmul_2510, conserved hypothetical protein; Bmul_2514, type 11 methyltransferase; wzt, ABC transporter ATP-binding protein; wzm, ABC transporter membrane permease; rmlDCAB, dTDP-rhamnose biosynthesis; wbxC, acetyltransferase; wbxD, glycosyltransferase; wbxE, glycosyltransferase; vioA, nucleotide sugar aminotransferase.