Figure 7.
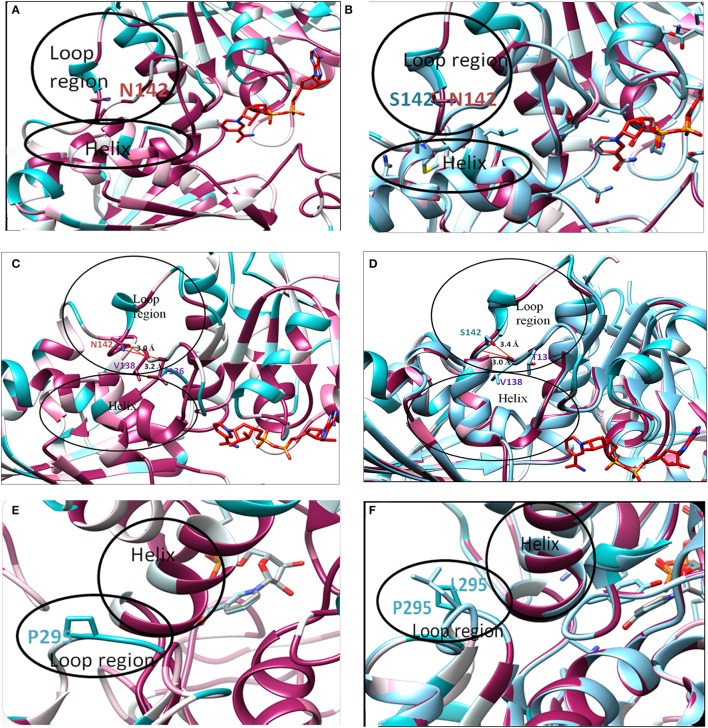
In silico analysis of mutations: Location of original and mutant residues (A) Model of Asparagine at position 142 of GAPDH (B) Structure showing the replacement of Asparagine with serine (C) Hydrogen bond formation is observed (represented as red line) between O-atom of Threonine 136 and NH2 group of Asparagine 142 (distance of interaction 3.2 Å). A second interaction occurs between the N-atom of Valine 138 and OH group of Asn 142 (distance of interaction 3.0 Å). (D) Formation of H-bond (represented as red line) is observed between O-atom of Valine 138 and N atom of Serine 142 (distance of interaction 3.0 Å) backbone interaction. A second interaction is observed between N-atom of Valine 138 and OH group of Serine 142 (distance of interaction 3.4 Å). As compared to the wild type Asparagine 142, the mutant Serine142 does not show any interaction with Threonine 136. (E) Model of Proline at position 295 of GAPDH (F) Structure showing the replacement of Proline with Leucine.