Abstract
Long interspersed nuclear element-1 (LINE-1) is a retrotransposon that contains a CpG island in its 5′-untranslated region. The CpG island of LINE-1 is often heavily methylated in normal somatic cells, which is associated with poor prognosis in various cancers. DNA methylation can differ between formalin-fixed paraffin-embedded (FFPE) and frozen tissues. Therefore, this study aimed to compare the LINE-1 methylation status between the two tissue-storage conditions in gastric cancer (GC) clinical samples and to evaluate whether LINE-1 can be used as an independent prognostic marker for each tissue-storage type. We analyzed four CpG sites of LINE-1 and examined the methylation levels at these sites in 25 FFPE and 41 frozen GC tissues by quantitative bisulfite pyrosequencing. The LINE-1 methylation status was significantly different between the FFPE and frozen GC tissues (p < 0.001). We further analyzed the clinicopathological features in the two groups separately. In the frozen GC tissues, LINE-1 was significantly hypomethylated in GC tissues compared to their corresponding normal gastric mucosa tissues (p < 0.001), and its methylation status was associated with gender, differentiation state, and lymphatic and venous invasion of GC. In the FFPE GC tissues, the methylation levels of LINE-1 differed according to tumor location and venous invasion of GC. In conclusion, LINE-1 can be used as a useful methylation marker for venous invasion in both FFPE and frozen tumor tissues of GC.
Keywords: gastric cancer, LINE-1, pyrosequencing, venous invasion
INTRODUCTION
Long interspersed nuclear element-1 (LINE-1) constitutes ~17% of the human genome (Lander et al., 2001). LINE-1 functions as a retrotransposon across its two open reading frames—ORF1p and ORF2p—and causes somatic retrotransposition, transcriptome effects, and DNA damage (Rodic and Burns, 2013). Thus far, many studies have proposed that the LINE-1 methylation status is associated with variant features in several cancer types. The hypomethylation of LINE-1, specifically, is associated with tumor progression, cancer risk, and poor prognosis in colon cancer, bladder cancer, breast cancer, and gastric cancer (GC) (Cash et al., 2012; Shigaki et al., 2013; Sunami et al., 2011; van Hoesel et al., 2012). LINE-1 is known to be hypomethylated in cancer tissues compared to normal tissues (Chalitchagorn et al., 2004). Previous studies reported that global LINE-1 hypomethylation was correlated with microsatellite instability and poor overall survival in colorectal cancer (Estecio et al., 2007; Ogino et al., 2008ba; 2008b). In addition, LINE-1 hypomethylation promotes liver metastasis by inducing expression of proto-oncogenes such as MET, RAB3IP, and CHRM 3 in colorectal cancer (Hur et al., 2014).
GC is the fifth most-common cancer in the world (Ferlay et al., 2015) and the second leading cause of cancer-related death in Asian countries (Rahman et al., 2014). The methylation status of LINE-1 differs between low-grade gastric dysplasia, high-grade dysplasia, and intramucosal cancer, and LINE-1 hypomethylation may be a diagnostic marker for high-grade dysplasia and intramucosal cancer (Lee et al., 2011). A previous study reported an association between LINE-1 hypomethylation and prognosis in gastric carcinogenesis (Shigaki et al., 2013). Moreover, the methylation status of LINE-1 changes when intestinal metaplasia progresses to gastric adenoma and is associated with poor prognosis in patients with GC (Bae et al., 2012). Thus, LINE-1 has been studied as an epigenetic marker associated with the risk for GC using blood or tissue samples. A meta-analysis of the LINE-1 methylation status in human cancers showed that LINE-1 is significantly hypomethylated as compared to controls in tissue samples such as fresh/frozen or formalin-fixed paraffin-embedded (FFPE) samples, but not in blood samples (Barchitta et al., 2014).
LINE-1 is associated with various clinicopathological features in cancer, particularly, tumor metastasis. In GC, the invading submucosa can cause lymph node (LN) metastasis. Although rarely observed, several patients have relatively high number of LN metastasis regardless of the depth of tumor invasion (T stage). While determining biomarkers for aggressive LN metastasis in GC, a previous study revealed that overexpressed plasminogen activator inhibitor-1 (PAI-1) is a promising marker for aggressive LN metastasis with low T stage (Suh et al., 2015). Furthermore, GCRG213p, a variant of LINE-1 endonuclease, is overexpressed in both primary GC and LN metastasis (Wang et al., 2013). Previously, LINE-1 methylation was analyzed at specific combined CpG sites using 434 FFPE tissues, to investigate associations with clinicopathological features, including lymphatic and venous invasion, and considering the tissue-storage conditions, LINE-1 was reportedly more hypermethylated in FFPE tissues than in paired fresh-frozen tissues in GC (Song et al., 2016).
Thus far, the relationship between the LINE-1 methylation level in different tissue types and several clinicopathological parameters such as differentiation, tumor location, lymphatic and venous invasion, and aggressive LN metastasis has not been investigated. Therefore, in the present study, we aimed to determine whether the LINE-1 methylation status could be used as a marker for GC in two tissue types, FFPE and frozen tissues. In addition, we assessed LINE-1 as an epigenetic marker for each tissue type and investigated the clinicopathological characteristics by LINE-1 methylation status in patients with GC.
MATERIALS AND METHODS
Tissue specimens
Forty-one frozen and 25 FFPE GC tissues were analyzed in this study. Pathological examination was performed by a pathologist to identify tumor cellularity or tumor area in the tissues. Frozen GC tissues were paired with normal gastric mucosa (NM), and only tumor area was used from FFPE tissues. The clinical characteristics of all patients are shown in Supplementary Tables S1 and S2.
All tissue samples were obtained from Seoul National University Hospital, Korea, and written informed consent was obtained from all patients. The present study was approved by the institutional review boards (IRB) of Seoul National University Hospital (IRB No. 1501-086-642).
LINE-1 methylation analysis
The analysis of LINE-1 methylation was performed in patients with GC who had data about gender, age, WHO classification, Lauren classification, tumor location, lymphatic invasion, venous invasion, perineural invasion, TNM stage and microsatellite instability. The methylation levels at four CpG sites of LINE-1 were measured in bisulfite-converted DNA from each tissue sample (Supplementary Fig. S1). To compare LINE-1 methylation levels, patients were grouped according to each clinicopathological parameter. In frozen tissues, different LINE-1 methylation levels in normal versus tumor tissue were also determined. In all the analysis, both the average methylation level of the four CpG sites and each CpG site were analyzed.
DNA extraction and sodium bisulfite modification
We collected tumor tissues from FFPE slides and placed them in microcentrifuge tubes with 1 ml xylene. After vortexing and centrifugation, the supernatant was removed. The pellet was washed twice with 1 ml 100% ethanol. Subsequently, genomic DNA (gDNA) was extracted using the QIAamp DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. GC tissues and the corresponding NM were immediately frozen in liquid nitrogen without fixation following gastrectomy. The gDNA was extracted from frozen tissues using the QuickGene DNA tissue kit S with QG-Mini80 (Kurabo, Japan) following the manufacturer’s instructions. Bisulfite-modified gDNA was prepared using the EpiTect Bisulfite Kit (Qiagen, Germany) according to the manufacturer’s instructions. The bisulfite reaction was carried out with 600 ng gDNA, and the reaction volume was adjusted to 40 μl with elution buffer. The sample tubes were placed in a thermal cycler (Eppendorf, Germany) under the following conditions: 5 min at 95°C, 25 min at 60°C, 5 min at 95°C, 85 min at 60°C, 5 min at 95°C, 175 min at 60°C, and overnight at 20°C. Converted DNA samples were finally stored at −20°C until further use.
Quantitative bisulfite pyrosequencing for LINE-1 methylation
We used the bisulfite pyrosequencing method for methylation analyses of the LINE-1 gene, as described previously (Kile et al., 2010). Polymerase chain reaction (PCR) was carried out in a reaction volume of 20 μl, with ≥20 ng converted gDNA, PCR premixture (Enzynomics, Korea), 1 μl of 10 pmol/μl forward primer, and 1 μl of 10 pmol/μl biotinylated-reverse primer. The amplification was carried out according to the general guidelines for pyrosequencing: denaturation at 95°C for 10 min; followed by 45 cycles at 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. The PCR products (2 μl) were separated by electrophoresis using a 2% agarose gel and visualized by ethidium bromide staining.
A single-stranded DNA template was prepared from 16–18 μl of the biotinylated PCR product using streptavidin Sepharose ® HP beads (Amersham Biosciences, Sweden) following the PSQ 96 sample preparation guide using multichannel pipettes. Fifteen picomoles of the respective sequencing primer were added for analysis. Sequencing was performed on a PyroMark ID system with a Pyro Gold reagents kit (Qiagen) according to the manufacturer’s instructions without further optimization. The methylation percentage was calculated from the average of the degree of methylation at 4 CpG sites formulated in pyrosequencing. The primer sequences are described in Supplementary Table S3.
Statistical analysis
A paired t-test was used to compare the methylation levels in GC tissues paired with NM. For the analyses of clinical implications, an unpaired t-test was used to analyze methylation levels among each classified groups according to clinicopathological parameters. The significant difference between samples considering the tumor location and TNM stage was calculated using the ANOVA test. Statistical significance was considered at p-value < 0.05. Statistical analyses were performed using GraphPad Prism V5.0 (GraphPad Software, USA)
RESULTS
LINE-1 methylation patterns according to sample types in GC
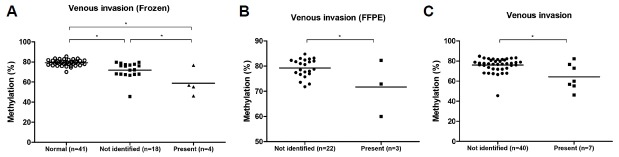
Comparison of the methylation levels at four CpG sites of the LINE-1 promoter showed that the average LINE-1 methylation was significantly different between the frozen and FFPE tumor tissues (Fig. 1A), and the differences were especially evident at CpG3 and CpG4 (Supplementary Fig. S2). Before we analyzed the clinical factors, we confirmed the LINE-1 methylation patterns at each CpG site according to the sample types. In frozen GC tissues, CpG3 of LINE-1 was significantly hypomethylated compared to other three CpG sites (CpG1; p = 0.031, CpG2; p < 0.001, CpG4; p < 0.001, Fig. 1B). In contrast, in FFPE GC tissues, CpG3 was significantly hypermethylated compared to CpG1 or CpG2 (CpG1; p = 0.022, CpG2; p < 0.001, Fig. 1C). In addition, CpG4 showed the maximum hypermethylation (Fig. 1C). Analysis of the LINE-1 methylation patterns according to the sample type and CpG sites showed that the LINE-1 methylation status varied between sample types and CpG sites in GC.
Fig. 1.
LINE-1 methylation status according to sample types.
(A) The average LINE-1 methylation level (%) was significantly different between the frozen tumor tissues and FFPE GC tissues (*p < 0.001, unpaired t test). (B) In 41 frozen GC tissues, LINE-1 methylation level (%) was significantly lower at CpG3 than at CpG1, CpG2 and CpG4 (p = 0.031, p < 0.001, p < 0.001, respectively). (C) In 25 FFPE GC tissues, CpG3 was significantly hypermethylated compared to CpG1 (p = 0.022) or CpG2 (p < 0.001). CpG4 showed the maximum hypermethylation compared to other three sites (p < 0.001, p < 0.001, p < 0.001, respectively). *p < 0.05, paired t test.
Association between LINE-1 methylation status and clinical characteristics in frozen GC tissues
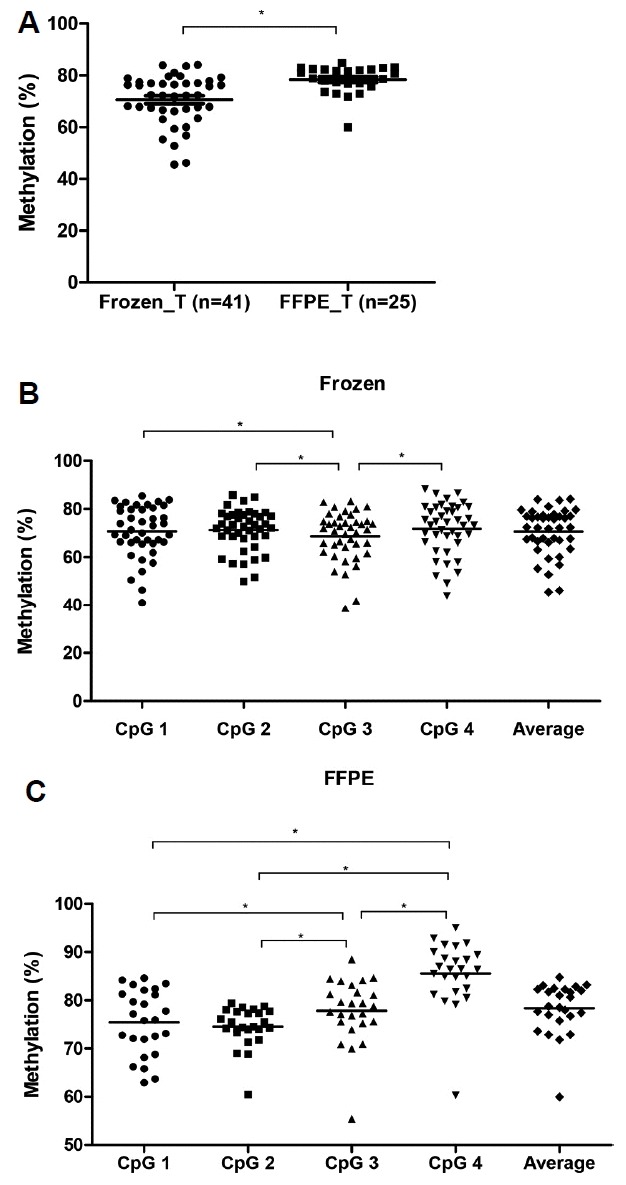
We analyzed LINE-1 methylation levels in each CpG site according to several clinicopathological characteristics by using frozen GC tissues, as shown in Table 1. LINE-1 at each CpG site showed hypomethylation in GC tissues compared to NM. Moreover, the LINE-1 methylation levels differed according to gender, WHO classification, lymphatic invasion, and venous invasion. Then, we also compared the average LINE-1 methylation level in each parameter. In the same manner, LINE-1 was significantly hypomethylated in tumor tissue compared to NM (Fig. 2A, p < 0.001), and the LINE-1 methylation level significantly differed between genders (Fig. 2B). Furthermore, we assessed LINE-1 methylation in GC tissue considering both differentiation and lymphatic invasion compared to NM. Although LINE-1 was significantly hypomethylated in GC irrespective of both the differentiation and lymphatic invasion statuses, it was more hypomethylated in differentiated gastric adenocarcinoma and in GC showing lymphatic invasion (Figs. 2C and 2D). Therefore, we believe that frozen GC tissues have a different LINE-1 methylation status in each differentiation and lymphatic invasion state and that LINE-1 hypomethylation is especially associated with the male gender, differentiated gastric adenocarcinoma, and the presence of lymphatic invasion.
Table 1.
LINE-1 methylation status in frozen GC tissues
| Characteristic | Number | LINE-1 methylation level (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| CpG 1 | P value | CpG 2 | P value | CpG 3 | P value | CpG 4 | P value | ||
| Type | |||||||||
| Normal mucosa | 41 | 81.61 | <0.001 | 78.27 | <0.001 | 76.32 | <0.001 | 80.48 | <0.001 |
| Gastric cancer | 41 | 70.73 | 71.32 | 68.74 | 71.76 | ||||
| Age | |||||||||
| ≤61 | 18 | 70.82 | 0.965 | 73.48 | 0.157 | 71.12 | 0.189 | 74.13 | 0.215 |
| >61 | 23 | 70.66 | 69.62 | 66.88 | 69.90 | ||||
| Gender | |||||||||
| Male | 30 | 68.42 | 0.021 | 69.19 | 0.007 | 65.69 | <0.001 | 69.12 | 0.008 |
| Female | 11 | 77.02 | 77.12 | 77.07 | 78.95 | ||||
| WHO classification | |||||||||
| Differentiated | 18 | 66.32 | 0.017 | 67.38 | 0.012 | 64.53 | 0.036 | 67.10 | 0.020 |
| Undifferentiated | 20 | 74.77 | 74.53 | 71.53 | 75.34 | ||||
| Lauren classification | |||||||||
| Intestinal | 20 | 67.18 | 0.199 | 68.40 | 0.252 | 65.25 | 0.211 | 68.14 | 0.214 |
| Diffuse | 12 | 72.53 | 71.97 | 70.02 | 73.12 | ||||
| Tumor location | |||||||||
| Upper | 11 | 69.59 | 0.887a | 68.70 | 0.295a | 65.29 | 0.296a | 68.12 | 0.207a |
| Middle | 8 | 69.85 | 75.02 | 72.80 | 77.04 | ||||
| Lower | 21 | 71.40 | 70.93 | 68.88 | 71.31 | ||||
| Lymphatic invasion | |||||||||
| Not identified | 11 | 75.53 | 0.041 | 74.28 | 0.020 | 72.00 | 0.020 | 75.35 | 0.029 |
| Present | 11 | 65.28 | 66.17 | 61.65 | 65.60 | ||||
| Venous invasion | |||||||||
| Not identified | 18 | 72.86 | 0.037 | 72.20 | 0.016 | 69.34 | 0.016 | 73.06 | 0.012 |
| Present | 4 | 59.36 | 61.36 | 55.50 | 58.82 | ||||
| Perineural invasion | |||||||||
| Not identified | 16 | 70.90 | 0.757 | 70.87 | 0.570 | 67.35 | 0.716 | 71.11 | 0.659 |
| Present | 6 | 69.07 | 68.51 | 65.41 | 68.77 | ||||
| TNM stage | |||||||||
| II | 26 | 70.92 | 0.922a | 70.59 | 0.535a | 67.58 | 0.487a | 71.01 | 0.532a |
| III | 13 | 69.98 | 71.79 | 70.01 | 72.00 | ||||
| IV | 2 | 73.10 | 77.64 | 75.73 | 79.98 | ||||
| Microsatellite instability | |||||||||
| MSS | 16 | 67.31 | 0.431 | 67.58 | 0.141 | 63.52 | 0.093 | 67.32 | 0.189 |
| MSI | 6 | 72.15 | 73.71 | 72.36 | 74.37 | ||||
ANOVA test
Fig. 2.
LINE-1 methylation and clinical characteristics in frozen GC tissues.
(A) LINE-1 was significantly hypomethylated in gastric tumor (T) compared to NM (N) (*p < 0.001, paired t test). (B) Significant difference of LINE-1 methylation level (%) between genders (p = 0.004). (C) LINE-1 was significantly hypomethylated in both differentiated and undifferentiated gastric adenocarcinoma compared to Normal (p < 0.001, p = 0.002, respectively). Particularly, it was more hypomethylated in differentiated. than in undifferentiated gastric adenocarcinoma. (p = 0.016). (D) Hypomethylated LINE-1 in tumor was observed irrespective of whether patients have lymphatic invasion or not (p < 0.001, p < 0.001, respectively). LINE-1 was significantly hypomethylated in the presence of invasion (Present) compared to the absence of it (Not identified) (p = 0.023). *p < 0.05 (unpaired t test).
Clinicopathological association of LINE-1 methylation status in FFPE GC tissues
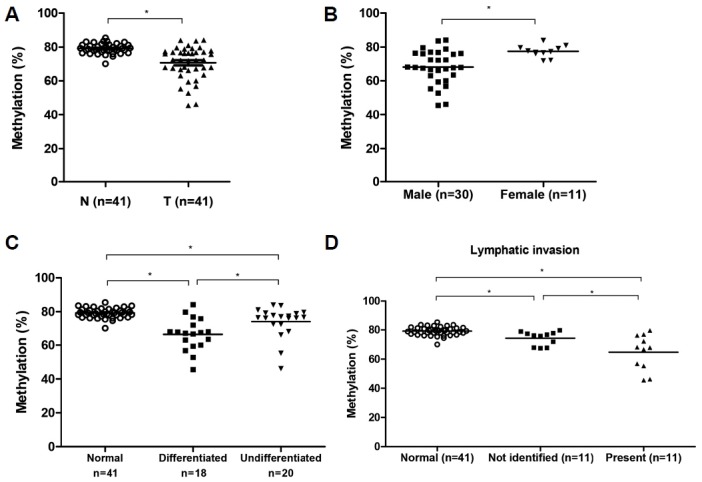
In the frozen GC tissues, LINE-1 methylation was associated with lymphatic invasion. Several patients had LN metastasis, although they were diagnosed with early GC. To determine whether LINE-1 methylation is a marker for aggressive LN metastasis, we obtained 25 FFPE tissue samples from patients with high T stage without LN metastasis (T3/4N0) or low T stage with >7 regional LN metastases (T1N3). However, we found no significant differences in the LINE-1 methylation levels between T3/4N0 and T1N3 (Fig. 3A). Therefore, we examined whether LINE-1 methylation in the FFPE tissues was associated with clinicopathological characteristics. We divided the patients into three groups according to tumor location (i.e., upper, middle, or lower third of the stomach), and on average, LINE-1 was significantly hypomethylated in tumors located in the lower third region compared to those in the middle third region (Fig. 3B). When we analyzed LINE-1 methylation patterns in each CpG site, the differences of LINE-1 methylation levels were also shown in several other characteristics (Table 2). On classifying patients into two groups according to the average age, only CpG1 of LINE-1 was significantly hypomethylated in patients older than 57 years. When tumor location was considered, only CpG1 and CpG2 were significantly different between tumors located in the middle and lower third regions (p = 0.002 and p = 0.008, respectively). In particular, LINE-1 methylation at CpG1 showed a significant difference according to the tumor location. The above-mentioned results indicate that LINE-1 hypomethylation occurs at CpG1 in aged patients with GC. In addition, LINE-1 is hypomethylated in tumors located in the lower third region considering FFPE GC tissues.
Fig. 3.
LINE-1 methylation and clinical characteristics in FFPE GC samples.
(A) No significant difference between two groups, high T stage without LN metastasis (T3/4N0) or low T stage with more than 7 regional LN metastasis (T1N3) (p = 0.211). (B) LINE-1 was significantly hypomethylated in tumors located in the lower third of stomach compared to those in the middle third of stomach (p = 0.008). *p < 0.05 (unpaired t test).
Table 2.
LINE-1 methylation status in FFPE GC tissues
| Characteristic | Number | LINE-1 methylation level (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| CpG 1 | P value | CpG 2 | P value | CpG 3 | P value | CpG 4 | P value | ||
| Type | |||||||||
| T3/4N0 | 13 | 73.78 | 0.207 | 73.64 | 0.250 | 76.84 | 0.446 | 83.98 | 0.226 |
| T1N3 | 12 | 77.26 | 75.57 | 78.91 | 87.31 | ||||
| Age | |||||||||
| ≤57 | 14 | 77.85 | 0.043 | 74.57 | 0.997 | 79.09 | 0.293 | 85.56 | 0.991 |
| >57 | 11 | 72.40 | 74.56 | 76.23 | 85.60 | ||||
| Gender | |||||||||
| Male | 12 | 75.25 | 0.889 | 74.23 | 0.707 | 76.88 | 0.500 | 85.16 | 0.774 |
| Female | 13 | 75.64 | 74.87 | 78.72 | 85.96 | ||||
| WHO classification | |||||||||
| Differentiated | 6 | 71.51 | 0.103 | 74.04 | 0.870 | 75.71 | 0.384 | 87.09 | 0.487 |
| Undifferentiated | 15 | 76.97 | 74.40 | 78.70 | 84.60 | ||||
| Lauren classification | |||||||||
| Intestinal | 8 | 71.58 | 0.090 | 74.28 | 0.955 | 76.07 | 0.518 | 87.35 | 0.300 |
| Diffuse | 15 | 76.55 | 74.18 | 77.96 | 84.14 | ||||
| Tumor location | |||||||||
| Upper | 5 | 75.44 | 0.014a | 73.59 | 0.156a | 75.17 | 0.252a | 82.11 | 0.367a |
| Middle | 5 | 82.75 | 0.002b | 77.73 | 0.008b | 81.95 | 0.080b | 88.19 | 0.319b |
| Lower | 15 | 73.02 | 73.83 | 77.35 | 85.87 | ||||
| Lymphatic invasion | |||||||||
| Not identified | 11 | 75.68 | 0.883 | 74.01 | 0.563 | 78.12 | 0.856 | 85.54 | 0.982 |
| Present | 14 | 75.27 | 75.00 | 77.62 | 85.61 | ||||
| Venous invasion | |||||||||
| Not identified | 22 | 76.11 | 0.189 | 75.17 | 0.042 | 79.10 | 0.007 | 86.67 | 0.025 |
| Present | 3 | 70.56 | 70.10 | 68.56 | 77.58 | ||||
| Perineural invasion | |||||||||
| Not identified | 12 | 77.65 | 0.120 | 75.24 | 0.440 | 78.52 | 0.631 | 85.69 | 0.939 |
| Present | 13 | 73.41 | 73.94 | 77.21 | 85.47 | ||||
| TNM stage | |||||||||
| II | 23 | 75.49 | 0.919 | 74.82 | 0.309 | 77.99 | 0.703 | 86.02 | 0.274 |
| IV | 2 | 74.97 | 71.67 | 76.07 | 80.47 | ||||
| Microsatellite instability | |||||||||
| MSS | 14 | 75.15 | 0.379 | 73.66 | 0.826 | 77.21 | 0.852 | 83.57 | 0.555 |
| MSI-high | 3 | 71.06 | 74.33 | 76.34 | 86.53 | ||||
ANOVA test
Unpaired t-test (middle vs. lower)
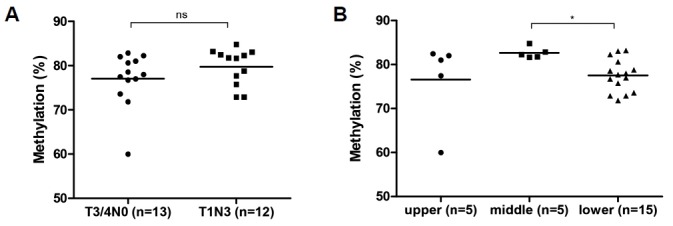
LINE-1 hypomethylation in the presence of venous invasion in frozen and FFPE tissues
When we analyzed the LINE-1 methylation according to lymphatic invasion, LINE-1 hypomethylation was associated with the presence of lymphatic invasion in frozen tissues but not in FFPE tissues. Further analysis using FFPE tissues did not show any significant difference between the absence of LN metastasis and the presence of aggressive LN metastasis. Therefore, we investigated whether LINE-1 methylation changes are associated with venous invasion in both frozen and FFPE tissues. LINE-1 in frozen tissues became hypomethylated in the presence of venous invasion compared to the absence of venous invasion or NM (Fig. 4A and Table 1). Moreover, in FFPE tissues, LINE-1 methylation significantly differed considering the presence and absence of venous invasion (Fig. 4B). The difference was significant at the CpG2, CpG3, and CpG4 sites (Table 2). In addition, we analyzed the average LINE-1 methylation level at CpG1 and CpG4 together according to all clinical characteristics, since a previous study showed that analysis of this combination was useful to predict prognosis for GC (Song et al., 2016). The results from this analysis were consistent with our findings from all CpG sites (data not shown), and combined two CpG sites was also a predictive factor of venous invasion (Supplementary Fig. S3). Therefore, LINE-1 hypomethylation was indicated in the presence of venous invasion in combined frozen and FFPE tissues (Fig. 4C). These results suggest that LINE-1 hypomethylation is strongly associated with venous invasion, irrespective of the tissue-storage type.
Fig. 4.
LINE-1 methylation and venous invasion in frozen and FFPE tissues.
(A) LINE-1 hypo-methylation in frozen GC was observed irrespective of whether patients have venous invasion or not (p < 0.001, p < 0.001, respectively). LINE-1 was significantly hypomethylated in the presence of invasion (Present) compared to the absence of it (Not identified) (p = 0.016). (B) LINE-1 methylation significantly differed considering the presence (Present) and absence of venous invasion (Not identified) (p = 0.017). (C) LINE-1 was significantly hypo-methylated in the presence of venous invasion in combined frozen and FFPE tissues (p = 0.001). *p < 0.05 (unpaired t test).
DISCUSSION
Considering the tissue-storage conditions in different cancer types, a previous study reported that colon tissues show clustered methylation patterns according to storage type (frozen vs. FFPE), and differentially methylated loci have been observed between the two sample types (Jasmine et al., 2012). Another study postulated that the methylation status of the MGMT promoter differs between paraffin-fixed and fresh-frozen tissues of glioblastoma (Hamilton et al., 2011). On evaluating 38 patients with GC, LINE-1 was reportedly more hypermethylated in FFPE tissues than in paired fresh-frozen tissues (Song et al., 2016).
Since the patient tissues were dissected for different purposes, they were preserved in various forms and given different pre-treatments. For pathological examination, GC tissues were fixed with formalin and embedded in paraffin. In contrast, for genomic analysis, tissues were immediately frozen in liquid nitrogen without fixation following gastrectomy. Bisulfite conversion can be affected by formalin fixation, as sequence artifacts such as cytosine deamination can be caused by formaldehyde (Do and Dobrovic, 2015). Therefore, we screened the LINE-1 methylation patterns in each tissue type separately to determine whether LINE-1 can be used as a marker under all circumstances. In the current study, we used samples from different patient groups, and different methylation levels were also observed between the two tissue types (Fig. 1A). Moreover, when we compared the LINE-1 methylation level in frozen GC tissues and paired adjacent NM, LINE-1 was significantly hypomethylated in GC tissue compared to NM (Fig. 2A), consistent with the findings of previous studies (Bae et al., 2012; Shigaki et al., 2013).
There are various methods to develop methylation biomarkers by quantification of DNA methylation. Recently, all routinely used assays for DNA methylation analysis were compared in different laboratories and countries. The results suggested that bisulfite pyrosequencing is a powerful and promising assay for developing biomarkers to measure absolute DNA methylation levels (Consortium, 2016). For LINE-1 methylation analysis, diverse assays, such as pyrosequencing, combined bisulfite restriction analysis, or MethyLight, have been used to measure methylation changes in samples from patients with cancer. However, among them, pyrosequencing is the most useful technique to detect LINE-1 methylation because it provides accurate and reliable results (Aparicio et al., 2009; Baba et al., 2014; Irahara et al., 2010). Therefore, this method is most commonly used in research on LINE-1 in human cancers.
Recently, Kim et al. reported that LINE-1 was differentially methylated according to the Vienna classification, and they suggested LINE-1 as a marker for early GC (Kim et al., 2016). Moreover, LINE-1 methylation at specific CpG sites was a prognostic marker in FFPE tissues obtained from 434 patients with advanced GC (Song et al., 2016). Likewise, several studies have evaluated LINE-1 methylation in GC, to the best of our knowledge, this is the first study to determine the association between LINE-1 methylation level and clinicopathological parameters in both frozen and FFPE sample types in GC. The current study had two main aims. The first was to determine the relationship between LINE-1 methylation and clinicopathological features using frozen tissues, and several features were associated with LINE-1 methylation change. The second aim was to analyze the relationship between LINE-1 methylation and aggressive LN metastasis using FFPE tissues, because we only obtained FFPE tissues from patients diagnosed with early GC (T1) with high LN metastasis (N3). However, we did not find any significant difference in LINE-1 methylation between T3/4N0 and T1N3 (Fig. 3A).
In other cancers, LINE-1 methylation was associated with venous invasion in various samples such as serum or frozen tissue (Ikeda et al., 2013; Tangkijvanich et al., 2007). In GC, vascular invasion can increase angiogenesis and distant metastasis including lymph node metastasis (Maehara et al., 2000). After curative resection, LINE-1 methylation is even associated with recurrence and poor survival in GC, even though patients do not have metastasis (Li et al., 2015). Accordingly, finding biomarkers for venous invasion can be a powerful tool to predict invasive potential and prognosis. Because LINE-1 methylation has been known to be associated with venous invasion in GC FFPE tissues, we additionally used frozen tissues to clarify whether LINE-1 could also be a biomarker for venous invasion in other sample types. LINE-1 hypomethylation was detected in the presence of venous invasion in both FFPE and frozen tissues (Fig. 4). Further study using blood samples is needed to investigate whether LINE-1 methylation can be a non-invasive marker in GC.
Despite our important findings, our present study has some limitations. First, we used FFPE GC tissues according to specific T and N stages without paired normal tissues to analyze the relationship between LINE-1 methylation and T1N3 GC. Therefore, we could not compare the LINE-1 methylation levels in both normal and GC tissues. Second, the sample size was relatively small. Although a significant difference in the LINE-1 methylation level was correlated with venous invasion in FFPE and frozen tissues, only few cases of venous invasion were analyzed. In further studies, a larger sample size should be included to increase the reliability and acquire statistically significant data. In particular, validation studies using larger sample sizes irrespective of the T or N stages should be conducted.
In conclusion, this study validated the LINE-1 methylation patterns in FFPE and frozen tissues in different GC patient groups and verified the relationships between clinicopathological parameters and LINE-1 methylation in each sample set. In frozen tissues, the LINE-1 methylation was associated with gender, differentiation state, and lymphatic and venous invasion. In FFPE tissues, the LINE-1 methylation levels significantly differed according to tumor location and venous invasion. Therefore, LINE-1 can be a useful marker for several clinical factors in each tissue-storage type and could be a biomarker for venous invasion in both frozen and FFPE GC tissues.
Supplementary data
ACKNOWLEDGMENTS
This work was supported by grants from the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant No. HI14C3426). This study was supported by the National Research Foundation of Korea (NRF) grant, funded by the Korean government (2014R1A5A2009242).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aparicio A., North B., Barske L., Wang X., Bollati V., Weisenberger D., Yoo C., Tannir N., Horne E., Groshen S., et al. LINE-1 methylation in plasma DNA as a biomarker of activity of DNA methylation inhibitors in patients with solid tumors. Epigenetics. 2009;4:176–184. doi: 10.4161/epi.4.3.8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y., Murata A., Watanabe M., Baba H. Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer. Surg Today. 2014;44:1807–1816. doi: 10.1007/s00595-013-0763-6. [DOI] [PubMed] [Google Scholar]
- Bae J.M., Shin S.H., Kwon H.J., Park S.Y., Kook M.C., Kim Y.W., Cho N.Y., Kim N., Kim T.Y., Kim D., et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int J Cancer. 2012;131:1323–1331. doi: 10.1002/ijc.27369. [DOI] [PubMed] [Google Scholar]
- Barchitta M., Quattrocchi A., Maugeri A., Vinciguerra M., Agodi A. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis. PLoS One. 2014;9:e109478. doi: 10.1371/journal.pone.0109478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash H.L., Tao L., Yuan J.M., Marsit C.J., Houseman E.A., Xiang Y.B., Gao Y.T., Nelson H.H., Kelsey K.T. LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. Int J Cancer. 2012;130:1151–1159. doi: 10.1002/ijc.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalitchagorn K., Shuangshoti S., Hourpai N., Kongruttanachok N., Tangkijvanich P., Thong-ngam D., Voravud N., Sriuranpong V., Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- Consortium B. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34:726–737. doi: 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- Do H., Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61:64–71. doi: 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- Estecio M.R., Gharibyan V., Shen L., Ibrahim A.E., Doshi K., He R., Jelinek J., Yang A.S., Yan P.S., Huang T.H., et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Hamilton M.G., Roldan G., Magliocco A., McIntyre J.B., Parney I., Easaw J.C. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol. 2011;102:255–260. doi: 10.1007/s11060-010-0307-5. [DOI] [PubMed] [Google Scholar]
- Hur K., Cejas P., Feliu J., Moreno-Rubio J., Burgos E., Boland C.R., Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–646. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Shiraishi K., Eguchi A., Shibata H., Yoshimoto K., Mori T., Baba Y., Baba H., Suzuki M. Long interspersed nucleotide element 1 hypomethylation is associated with poor prognosis of lung adenocarcinoma. Ann Thorac Surg. 2013;96:1790–1794. doi: 10.1016/j.athoracsur.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Irahara N., Nosho K., Baba Y., Shima K., Lindeman N.I., Hazra A., Schernhammer E.S., Hunter D.J., Fuchs C.S., Ogino S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmine F., Rahaman R., Roy S., Raza M., Paul R., Rakibuz-Zaman M., Paul-Brutus R., Dodsworth C., Kamal M., Ahsan H., et al. Interpretation of genome-wide infinium methylation data from ligated DNA in formalin-fixed, paraffin-embedded paired tumor and normal tissue. BMC Res Notes. 2012;5:117. doi: 10.1186/1756-0500-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile M.L., Baccarelli A., Tarantini L., Hoffman E., Wright R.O., Christiani D.C. Correlation of global and gene-specific DNA methylation in maternal-infant pairs. PLoS One. 2010;5:e13730. doi: 10.1371/journal.pone.0013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.J., Chung W.C., Kim D.B., Kim Y.J., Lee J.M., Jung J.H., Lee Y.K. Long interspersed nuclear element (LINE)-1 methylation level as a molecular marker of early gastric cancer. Dig Liver Dis. 2016;48:1093–1097. doi: 10.1016/j.dld.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee J.R., Chung W.C., Kim J.D., Lee K.M., Paik C.N., Jung S.H., Jung J.H., Lee Y.K., Han S.W. Differential LINE-1 hypomethylation of gastric low-grade dysplasia from high grade dysplasia and intramucosal cancer. Gut Liver. 2011;5:149–153. doi: 10.5009/gnl.2011.5.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ling Y.H., Zhu C.M., Hu W.M., Zhang X.K., Luo R.Z., He J.H., Yun J.P., Li Y.F., Cai M.Y. Vascular invasion as an independent predictor of poor prognosis in nonmetastatic gastric cancer after curative resection. Int J Clin Exp Pathol. 2015;8:3910–3918. [PMC free article] [PubMed] [Google Scholar]
- Maehara Y., Kabashima A., Koga T., Tokunaga E., Takeuchi H., Kakeji Y., Sugimachi K. Vascular invasion and potential for tumor angiogenesis and metastasis in gastric carcinoma. Surgery. 2000;128:408–416. doi: 10.1067/msy.2000.107265. [DOI] [PubMed] [Google Scholar]
- Ogino S., Kawasaki T., Nosho K., Ohnishi M., Suemoto Y., Kirkner G.J., Fuchs C.S. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008a;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S., Nosho K., Kirkner G.J., Kawasaki T., Chan A.T., Schernhammer E.S., Giovannucci E.L., Fuchs C.S. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008b;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R., Asombang A.W., Ibdah J.A. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20:4483–4490. doi: 10.3748/wjg.v20.i16.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodic N., Burns K.H. Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet. 2013;9:e1003402. doi: 10.1371/journal.pgen.1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki H., Baba Y., Watanabe M., Murata A., Iwagami S., Miyake K., Ishimoto T., Iwatsuki M., Baba H. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16:480–487. doi: 10.1007/s10120-012-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.S., Kim Y., Cho N.Y., Yang H.K., Kim W.H., Kang G.H. Methylation status of long interspersed element-1 in advanced gastric cancer and its prognostic implication. Gastric Cancer. 2016;19:98–106. doi: 10.1007/s10120-015-0463-6. [DOI] [PubMed] [Google Scholar]
- Suh Y.S., Yu J., Kim B.C., Choi B., Han T.S., Ahn H.S., Kong S.H., Lee H.J., Kim W.H., Yang H.K. Overexpression of plasminogen activator inhibitor-1 in advanced gastric cancer with aggressive lymph node metastasis. Cancer Res Treat. 2015;47:718–726. doi: 10.4143/crt.2014.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunami E., de Maat M., Vu A., Turner R.R., Hoon D.S. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangkijvanich P., Hourpai N., Rattanatanyong P., Wisedopas N., Mahachai V., Mutirangura A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta. 2007;379:127–133. doi: 10.1016/j.cca.2006.12.029. [DOI] [PubMed] [Google Scholar]
- van Hoesel A.Q., van de Velde C.J., Kuppen P.J., Liefers G.J., Putter H., Sato Y., Elashoff D.A., Turner R.R., Shamonki J.M., de Kruijf E.M., et al. Hypomethylation of LINE-1 in primary tumor has poor prognosis in young breast cancer patients: a retrospective cohort study. Breast Cancer Res Treat. 2012;134:1103–1114. doi: 10.1007/s10549-012-2038-0. [DOI] [PubMed] [Google Scholar]
- Wang G., Gao J., Huang H., Tian Y., Xue L., Wang W., You W., Lian H., Duan X., Wu B., et al. Expression of a LINE-1 endonuclease variant in gastric cancer: its association with clinicopathological parameters. BMC Cancer. 2013;13:265. doi: 10.1186/1471-2407-13-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.