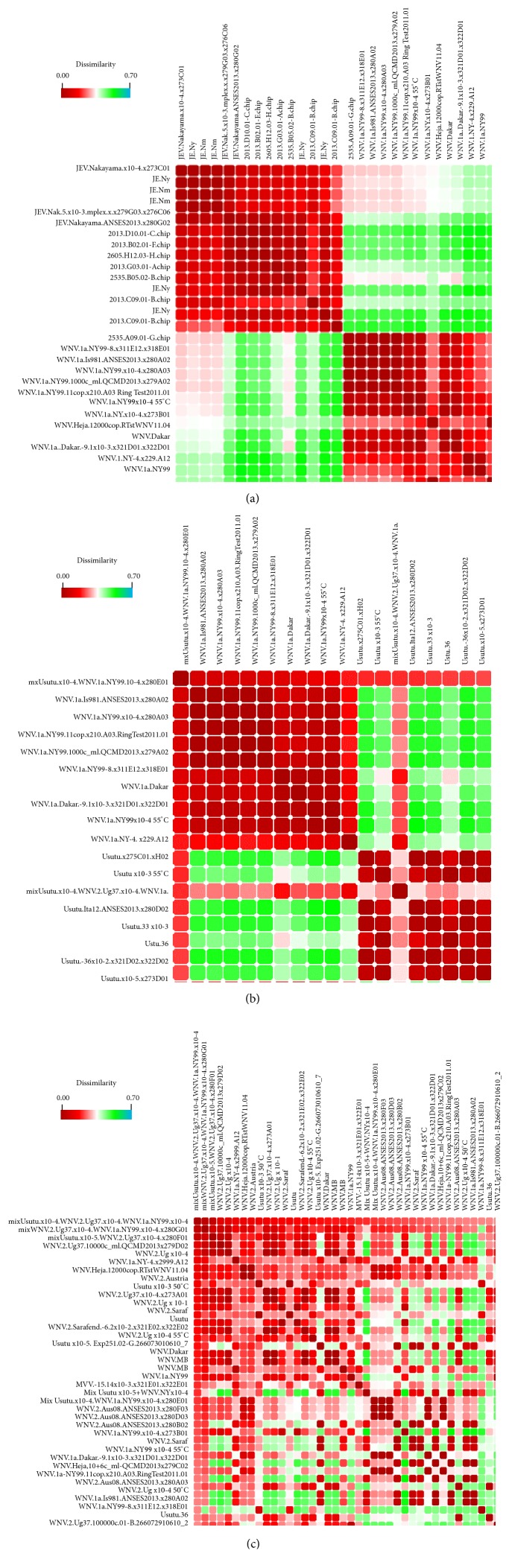
Figure 5.
Presentation of experimental results processed by the Orange's heat-map-like widget showing an all-versus-all sample comparison. Calculated distances between intensity patterns of each pair of microarray results obtained from hybridization of sample amplicons are shown, with each cell representing the comparison of two microarray experiments. The dissimilarity color scale is shown at the top. Columns and rows are organized in the same order, making the diagonal an “identity” sample comparison. When one sample is selected by mouse-clicking, the map is immediately reorganized to show this sample in the upper row and left column followed by the most similar samples. (a) The selection of the “JEV.Nakayama.x10-4.x273C01” sample reveals a good separation of JEV samples from all other viral species. (b) The same heat map after selecting a sample containing a mix of diluted USUV and WNV1a RNA, which is easily recognized by a framed mosaic-like map. (c) A more complex mix, including USUV, WNV1, and WNV-2 RNA, was selected showing a more complex pattern that still permits the identification of the components (which can be additionally confirmed by the successive selection of each of the three standard WNV-1, WNV-2, and USUV to check that this mix clusters together with each of them).