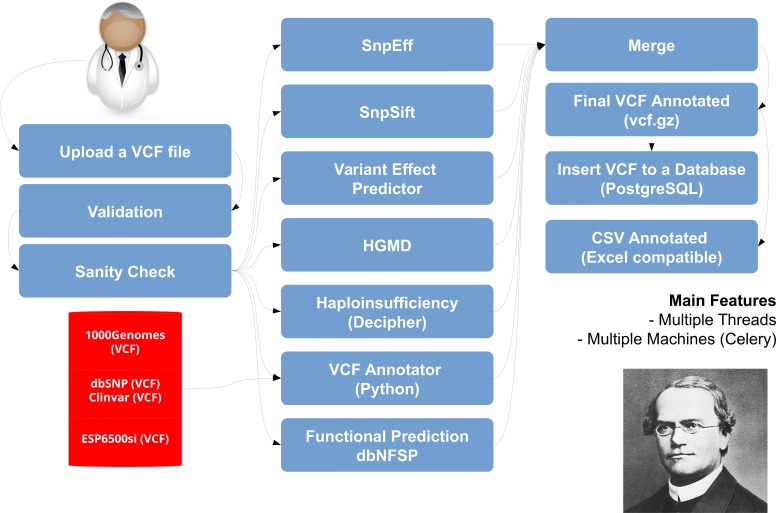
Fig 2. Pynnotator annotation framework.
This figure shows the processes we perform on each VCF file after a user uploads it to the system. First we perform a vcf-validation and a sanity check to make sure the file is ready to be annotated. After it, we send each checked VCF file to be annotated in parallel by SNPEFF, SNPSIFT, VEP, Decipher, and two other python scripts we developed in-house. The first one is capable of using other VCF files to annotate a VCF file so we use the VCFs from 1000Genomes, dbSNP and NHLBI GO Exome Sequencing Project to annotate a VCF and the second python script uses data from dbNSFP to add functional annotation and prediction scores to our annotation. After merging the results of all tasks we compress the file and prepare it to be inserted to the database.