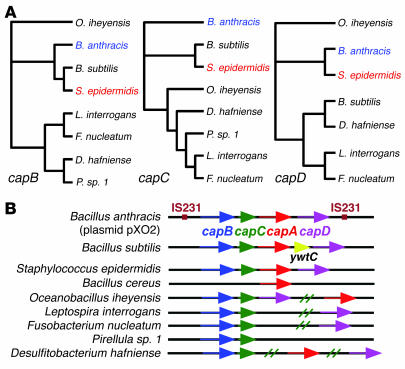
Figure 1.
Molecular genetic comparison of bacteria with genes encoding a putative PGA synthesis machinery. (A) Phylogenetic trees based on sequence comparisons of the capB (amide ligase), capC (unknown function), and capD (depolymerase) genes. CapA is a putative PGA exporter. We have excluded a comparison of capA genes, because capA homologs were not found in all the organisms and comparison of transporters is normally less indicative of phylogenetic relations. Of the microorganisms shown, production of PGA has been demonstrated previously only in B. anthracis and B. subtilis, and in this study, in S. epidermidis. (B) cap genes and homologs in bacteria for which genetic information is available. The B. anthracis cap gene cluster is located on a plasmid and flanked by IS231 insertion sequences. All other genes are located in the bacterial chromosomes.